
Pustulibacterium marinum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Pustulibacterium
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
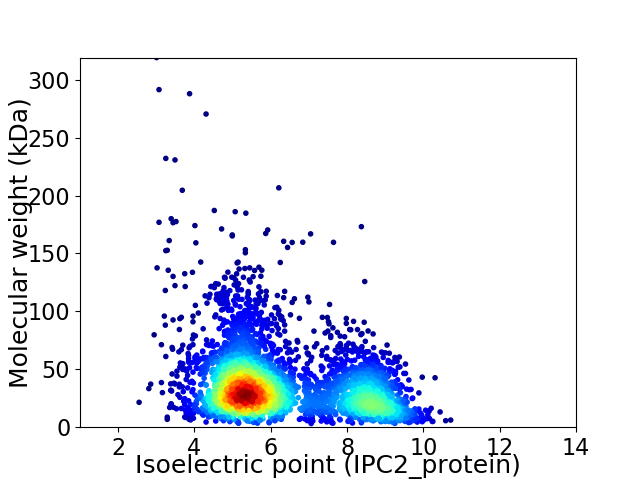
Virtual 2D-PAGE plot for 3719 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I7FBT0|A0A1I7FBT0_9FLAO ZIP Zinc transporter OS=Pustulibacterium marinum OX=1224947 GN=SAMN05216480_101826 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.44KK3 pKa = 8.49ITFLFLLVASFTLNAQLVVNEE24 pKa = 4.41VDD26 pKa = 3.34ADD28 pKa = 4.09TPGVDD33 pKa = 3.01NQEE36 pKa = 4.32FIEE39 pKa = 4.67LKK41 pKa = 10.49SDD43 pKa = 3.73TPNLSLDD50 pKa = 3.69GYY52 pKa = 10.47VVVLFNGNGDD62 pKa = 3.47YY63 pKa = 10.8SYY65 pKa = 11.75YY66 pKa = 11.0SVDD69 pKa = 3.54LDD71 pKa = 5.28GYY73 pKa = 7.38TTDD76 pKa = 4.01DD77 pKa = 3.41NGIIVIGSAGVSPFPGILISANVIQNGADD106 pKa = 2.96AVAIYY111 pKa = 9.99QGSWYY116 pKa = 9.58DD117 pKa = 3.8YY118 pKa = 11.24PEE120 pKa = 4.55DD121 pKa = 4.01TPPTTEE127 pKa = 3.89NLIDD131 pKa = 3.85ALVYY135 pKa = 8.54DD136 pKa = 4.92TNDD139 pKa = 3.25SDD141 pKa = 4.91DD142 pKa = 3.98TGLMAALGVTTQTNEE157 pKa = 3.78GGNGSTTSNSVQRR170 pKa = 11.84MNDD173 pKa = 3.06GTYY176 pKa = 10.91VSATPTPGLPNDD188 pKa = 4.04GSGVVYY194 pKa = 10.91NGITAQLSANEE205 pKa = 4.07FVEE208 pKa = 4.46GDD210 pKa = 3.7SFSITFTTEE219 pKa = 3.73DD220 pKa = 3.8NVDD223 pKa = 3.32EE224 pKa = 4.43TLVFDD229 pKa = 4.97ISFDD233 pKa = 3.68NEE235 pKa = 4.14GFSSEE240 pKa = 4.82DD241 pKa = 2.94ISGNFSISIASGTSSVTEE259 pKa = 4.42TYY261 pKa = 10.63QIIDD265 pKa = 3.67DD266 pKa = 4.37TEE268 pKa = 5.33DD269 pKa = 3.19EE270 pKa = 4.26GDD272 pKa = 3.68EE273 pKa = 4.34VFVLNMINIPAGFNLLNNLLEE294 pKa = 4.25GTITDD299 pKa = 4.24DD300 pKa = 5.63DD301 pKa = 5.16YY302 pKa = 11.91STSAWGTPLNPTYY315 pKa = 10.91GIVSSTAPDD324 pKa = 3.32NYY326 pKa = 10.49YY327 pKa = 11.12SSLDD331 pKa = 3.81GLSGAALEE339 pKa = 4.78QAIQDD344 pKa = 4.34IIADD348 pKa = 4.03PNVVRR353 pKa = 11.84AQNYY357 pKa = 10.39GDD359 pKa = 3.41VTDD362 pKa = 4.63ILKK365 pKa = 10.69QSDD368 pKa = 3.72QNPEE372 pKa = 3.8NNNEE376 pKa = 3.21VWLIYY381 pKa = 9.81TEE383 pKa = 4.09QPRR386 pKa = 11.84AKK388 pKa = 9.47IDD390 pKa = 3.6FQTTSNNTGTWNRR403 pKa = 11.84EE404 pKa = 3.61HH405 pKa = 7.71IYY407 pKa = 9.15PQSRR411 pKa = 11.84GGFSGGTEE419 pKa = 4.05YY420 pKa = 10.75EE421 pKa = 3.67ADD423 pKa = 4.67GIDD426 pKa = 4.16VYY428 pKa = 11.55LPTNADD434 pKa = 4.25DD435 pKa = 4.03ILAGHH440 pKa = 7.38ADD442 pKa = 3.26AHH444 pKa = 6.7HH445 pKa = 6.54IRR447 pKa = 11.84AVDD450 pKa = 3.84GPEE453 pKa = 3.56NSSRR457 pKa = 11.84GNQDD461 pKa = 3.17YY462 pKa = 11.25GSGAYY467 pKa = 8.64EE468 pKa = 4.43GPTANAGSWKK478 pKa = 10.43GDD480 pKa = 3.25VARR483 pKa = 11.84SLFYY487 pKa = 10.25MAIRR491 pKa = 11.84YY492 pKa = 8.7NDD494 pKa = 3.7LDD496 pKa = 4.01VVNGNPNDD504 pKa = 3.78STVGQLGDD512 pKa = 3.78LATLLVWNTQDD523 pKa = 3.64PADD526 pKa = 4.13DD527 pKa = 4.26FEE529 pKa = 4.99MNRR532 pKa = 11.84NNYY535 pKa = 9.06IYY537 pKa = 9.19TWQYY541 pKa = 9.7NRR543 pKa = 11.84NPFIDD548 pKa = 4.03YY549 pKa = 10.39PEE551 pKa = 4.22LADD554 pKa = 5.65YY555 pKa = 10.68IWGDD559 pKa = 3.33NAGEE563 pKa = 4.24VWNLPMSTHH572 pKa = 5.68QEE574 pKa = 3.28ILEE577 pKa = 4.11AVKK580 pKa = 10.17IYY582 pKa = 9.57PNPINSYY589 pKa = 8.94FQITGISEE597 pKa = 4.07EE598 pKa = 4.14FKK600 pKa = 10.02ITIMDD605 pKa = 3.66STGKK609 pKa = 10.17VVLSKK614 pKa = 11.11SDD616 pKa = 3.08VDD618 pKa = 4.36SNRR621 pKa = 11.84KK622 pKa = 9.29IYY624 pKa = 10.18IDD626 pKa = 4.04GSAGIYY632 pKa = 10.2LVTIEE637 pKa = 4.53TEE639 pKa = 3.46KK640 pKa = 10.37GTIRR644 pKa = 11.84KK645 pKa = 9.45KK646 pKa = 9.76IIKK649 pKa = 8.5MM650 pKa = 3.59
MM1 pKa = 7.45KK2 pKa = 10.44KK3 pKa = 8.49ITFLFLLVASFTLNAQLVVNEE24 pKa = 4.41VDD26 pKa = 3.34ADD28 pKa = 4.09TPGVDD33 pKa = 3.01NQEE36 pKa = 4.32FIEE39 pKa = 4.67LKK41 pKa = 10.49SDD43 pKa = 3.73TPNLSLDD50 pKa = 3.69GYY52 pKa = 10.47VVVLFNGNGDD62 pKa = 3.47YY63 pKa = 10.8SYY65 pKa = 11.75YY66 pKa = 11.0SVDD69 pKa = 3.54LDD71 pKa = 5.28GYY73 pKa = 7.38TTDD76 pKa = 4.01DD77 pKa = 3.41NGIIVIGSAGVSPFPGILISANVIQNGADD106 pKa = 2.96AVAIYY111 pKa = 9.99QGSWYY116 pKa = 9.58DD117 pKa = 3.8YY118 pKa = 11.24PEE120 pKa = 4.55DD121 pKa = 4.01TPPTTEE127 pKa = 3.89NLIDD131 pKa = 3.85ALVYY135 pKa = 8.54DD136 pKa = 4.92TNDD139 pKa = 3.25SDD141 pKa = 4.91DD142 pKa = 3.98TGLMAALGVTTQTNEE157 pKa = 3.78GGNGSTTSNSVQRR170 pKa = 11.84MNDD173 pKa = 3.06GTYY176 pKa = 10.91VSATPTPGLPNDD188 pKa = 4.04GSGVVYY194 pKa = 10.91NGITAQLSANEE205 pKa = 4.07FVEE208 pKa = 4.46GDD210 pKa = 3.7SFSITFTTEE219 pKa = 3.73DD220 pKa = 3.8NVDD223 pKa = 3.32EE224 pKa = 4.43TLVFDD229 pKa = 4.97ISFDD233 pKa = 3.68NEE235 pKa = 4.14GFSSEE240 pKa = 4.82DD241 pKa = 2.94ISGNFSISIASGTSSVTEE259 pKa = 4.42TYY261 pKa = 10.63QIIDD265 pKa = 3.67DD266 pKa = 4.37TEE268 pKa = 5.33DD269 pKa = 3.19EE270 pKa = 4.26GDD272 pKa = 3.68EE273 pKa = 4.34VFVLNMINIPAGFNLLNNLLEE294 pKa = 4.25GTITDD299 pKa = 4.24DD300 pKa = 5.63DD301 pKa = 5.16YY302 pKa = 11.91STSAWGTPLNPTYY315 pKa = 10.91GIVSSTAPDD324 pKa = 3.32NYY326 pKa = 10.49YY327 pKa = 11.12SSLDD331 pKa = 3.81GLSGAALEE339 pKa = 4.78QAIQDD344 pKa = 4.34IIADD348 pKa = 4.03PNVVRR353 pKa = 11.84AQNYY357 pKa = 10.39GDD359 pKa = 3.41VTDD362 pKa = 4.63ILKK365 pKa = 10.69QSDD368 pKa = 3.72QNPEE372 pKa = 3.8NNNEE376 pKa = 3.21VWLIYY381 pKa = 9.81TEE383 pKa = 4.09QPRR386 pKa = 11.84AKK388 pKa = 9.47IDD390 pKa = 3.6FQTTSNNTGTWNRR403 pKa = 11.84EE404 pKa = 3.61HH405 pKa = 7.71IYY407 pKa = 9.15PQSRR411 pKa = 11.84GGFSGGTEE419 pKa = 4.05YY420 pKa = 10.75EE421 pKa = 3.67ADD423 pKa = 4.67GIDD426 pKa = 4.16VYY428 pKa = 11.55LPTNADD434 pKa = 4.25DD435 pKa = 4.03ILAGHH440 pKa = 7.38ADD442 pKa = 3.26AHH444 pKa = 6.7HH445 pKa = 6.54IRR447 pKa = 11.84AVDD450 pKa = 3.84GPEE453 pKa = 3.56NSSRR457 pKa = 11.84GNQDD461 pKa = 3.17YY462 pKa = 11.25GSGAYY467 pKa = 8.64EE468 pKa = 4.43GPTANAGSWKK478 pKa = 10.43GDD480 pKa = 3.25VARR483 pKa = 11.84SLFYY487 pKa = 10.25MAIRR491 pKa = 11.84YY492 pKa = 8.7NDD494 pKa = 3.7LDD496 pKa = 4.01VVNGNPNDD504 pKa = 3.78STVGQLGDD512 pKa = 3.78LATLLVWNTQDD523 pKa = 3.64PADD526 pKa = 4.13DD527 pKa = 4.26FEE529 pKa = 4.99MNRR532 pKa = 11.84NNYY535 pKa = 9.06IYY537 pKa = 9.19TWQYY541 pKa = 9.7NRR543 pKa = 11.84NPFIDD548 pKa = 4.03YY549 pKa = 10.39PEE551 pKa = 4.22LADD554 pKa = 5.65YY555 pKa = 10.68IWGDD559 pKa = 3.33NAGEE563 pKa = 4.24VWNLPMSTHH572 pKa = 5.68QEE574 pKa = 3.28ILEE577 pKa = 4.11AVKK580 pKa = 10.17IYY582 pKa = 9.57PNPINSYY589 pKa = 8.94FQITGISEE597 pKa = 4.07EE598 pKa = 4.14FKK600 pKa = 10.02ITIMDD605 pKa = 3.66STGKK609 pKa = 10.17VVLSKK614 pKa = 11.11SDD616 pKa = 3.08VDD618 pKa = 4.36SNRR621 pKa = 11.84KK622 pKa = 9.29IYY624 pKa = 10.18IDD626 pKa = 4.04GSAGIYY632 pKa = 10.2LVTIEE637 pKa = 4.53TEE639 pKa = 3.46KK640 pKa = 10.37GTIRR644 pKa = 11.84KK645 pKa = 9.45KK646 pKa = 9.76IIKK649 pKa = 8.5MM650 pKa = 3.59
Molecular weight: 70.82 kDa
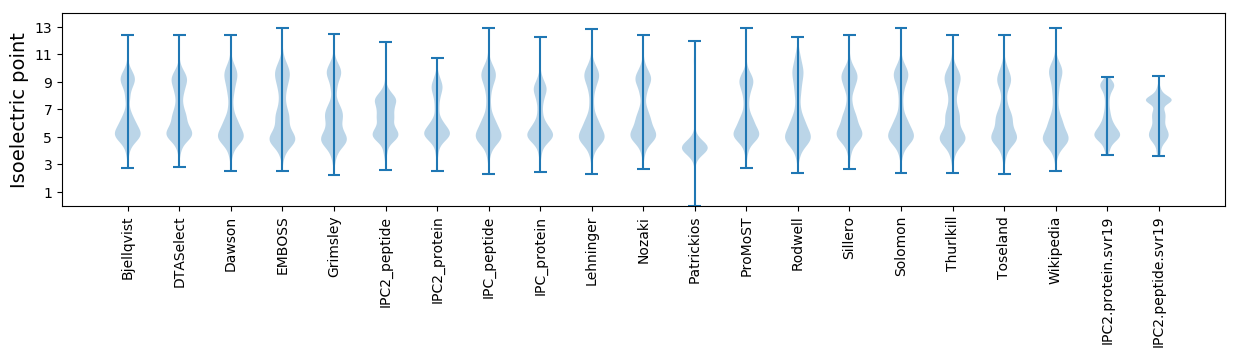
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I7G5E1|A0A1I7G5E1_9FLAO OstA-like protein OS=Pustulibacterium marinum OX=1224947 GN=SAMN05216480_103196 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.12VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.34KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.12VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.34KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
Molecular weight: 6.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1275365 |
29 |
2940 |
342.9 |
38.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.676 ± 0.038 | 0.773 ± 0.014 |
5.615 ± 0.037 | 6.898 ± 0.037 |
5.163 ± 0.034 | 6.303 ± 0.038 |
1.85 ± 0.018 | 7.422 ± 0.041 |
7.323 ± 0.05 | 9.032 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.019 | 5.882 ± 0.041 |
3.394 ± 0.022 | 3.666 ± 0.026 |
3.186 ± 0.027 | 6.394 ± 0.031 |
6.231 ± 0.045 | 6.365 ± 0.034 |
1.121 ± 0.015 | 4.407 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
