
Rhodosporidium toruloides (strain ATCC 204091 / IIP 30 / MTCC 1151) (Yeast) (Rhodotorula glutinis (strain ATCC 204091))
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Pucciniomycotina; Microbotryomycetes; Sporidiobolales; Sporidiobolaceae; Rhodotorula; Rhodotorula toruloides
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
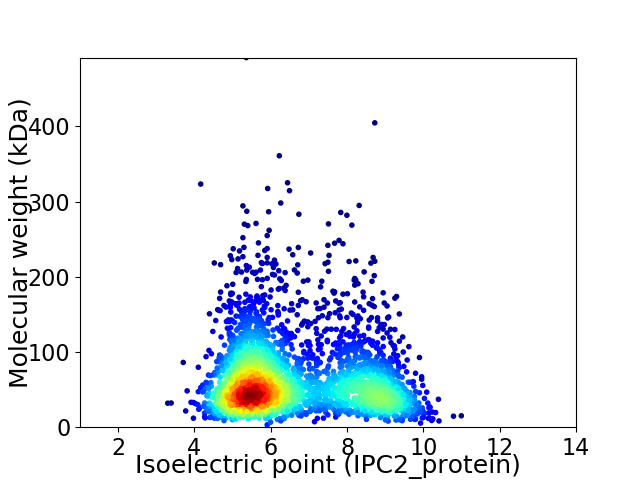
Virtual 2D-PAGE plot for 2816 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0SUL3|G0SUL3_RHOT2 Uncharacterized protein OS=Rhodosporidium toruloides (strain ATCC 204091 / IIP 30 / MTCC 1151) OX=1001064 GN=RTG_00124 PE=4 SV=1
MM1 pKa = 7.52LLYY4 pKa = 9.56ATLAVIFACSGLVSAAPATAAAGGTASSLDD34 pKa = 3.54ASSKK38 pKa = 10.8AFAAAVSYY46 pKa = 11.3QEE48 pKa = 4.03TCYY51 pKa = 10.21LTLFDD56 pKa = 5.39ADD58 pKa = 4.02EE59 pKa = 4.72NGQDD63 pKa = 3.47VYY65 pKa = 11.51INDD68 pKa = 4.47GYY70 pKa = 11.55KK71 pKa = 9.82KK72 pKa = 10.22DD73 pKa = 3.86SSGKK77 pKa = 8.24EE78 pKa = 3.8YY79 pKa = 10.68YY80 pKa = 10.11YY81 pKa = 10.95EE82 pKa = 4.0EE83 pKa = 4.44ACYY86 pKa = 10.3KK87 pKa = 10.26SSKK90 pKa = 10.38VDD92 pKa = 4.26GKK94 pKa = 7.91TQSNKK99 pKa = 9.3VASSSTTDD107 pKa = 3.2NSPFSSLIGTLGVDD121 pKa = 3.7DD122 pKa = 4.68SVSGFLSGLVSGDD135 pKa = 3.5SLGLGGLVRR144 pKa = 11.84FVKK147 pKa = 10.43RR148 pKa = 11.84QFDD151 pKa = 3.78GKK153 pKa = 9.36TIGAILGQPAAEE165 pKa = 4.29PAASSSSSAAAPTPEE180 pKa = 4.8EE181 pKa = 4.84DD182 pKa = 3.54SATPSSPDD190 pKa = 3.3YY191 pKa = 10.92FDD193 pKa = 5.23APSPTPTPIPTTTAAPSPTAVNPLEE218 pKa = 4.17IANGFGGIQSVGNALPGRR236 pKa = 11.84IGAEE240 pKa = 4.37DD241 pKa = 3.32ILGPALSSANADD253 pKa = 3.41PTSAAFGATTTDD265 pKa = 2.8AGVPATQAAPQQSEE279 pKa = 4.48TPDD282 pKa = 3.64AQPSTSAFTPAPGPPGYY299 pKa = 7.79PTAFTPAAGPPSSPSSFTPADD320 pKa = 4.13GPPSASPTVALNPNNVDD337 pKa = 3.12NAAPQATQFQPVSSPPVVQAPASTFTPAAAPSTQDD372 pKa = 3.14VASPATTQQIQPTPSSLVVQTAAPPVNSAPSFFTSASGVVTPTILATSSPVAADD426 pKa = 3.74TPTPAATSAPSSTVVDD442 pKa = 4.71TPATSSTVPLRR453 pKa = 11.84IYY455 pKa = 10.17VAAASPSSSSVDD467 pKa = 3.18AASSTTTPLTLVSLAPTAVSAQSAA491 pKa = 3.6
MM1 pKa = 7.52LLYY4 pKa = 9.56ATLAVIFACSGLVSAAPATAAAGGTASSLDD34 pKa = 3.54ASSKK38 pKa = 10.8AFAAAVSYY46 pKa = 11.3QEE48 pKa = 4.03TCYY51 pKa = 10.21LTLFDD56 pKa = 5.39ADD58 pKa = 4.02EE59 pKa = 4.72NGQDD63 pKa = 3.47VYY65 pKa = 11.51INDD68 pKa = 4.47GYY70 pKa = 11.55KK71 pKa = 9.82KK72 pKa = 10.22DD73 pKa = 3.86SSGKK77 pKa = 8.24EE78 pKa = 3.8YY79 pKa = 10.68YY80 pKa = 10.11YY81 pKa = 10.95EE82 pKa = 4.0EE83 pKa = 4.44ACYY86 pKa = 10.3KK87 pKa = 10.26SSKK90 pKa = 10.38VDD92 pKa = 4.26GKK94 pKa = 7.91TQSNKK99 pKa = 9.3VASSSTTDD107 pKa = 3.2NSPFSSLIGTLGVDD121 pKa = 3.7DD122 pKa = 4.68SVSGFLSGLVSGDD135 pKa = 3.5SLGLGGLVRR144 pKa = 11.84FVKK147 pKa = 10.43RR148 pKa = 11.84QFDD151 pKa = 3.78GKK153 pKa = 9.36TIGAILGQPAAEE165 pKa = 4.29PAASSSSSAAAPTPEE180 pKa = 4.8EE181 pKa = 4.84DD182 pKa = 3.54SATPSSPDD190 pKa = 3.3YY191 pKa = 10.92FDD193 pKa = 5.23APSPTPTPIPTTTAAPSPTAVNPLEE218 pKa = 4.17IANGFGGIQSVGNALPGRR236 pKa = 11.84IGAEE240 pKa = 4.37DD241 pKa = 3.32ILGPALSSANADD253 pKa = 3.41PTSAAFGATTTDD265 pKa = 2.8AGVPATQAAPQQSEE279 pKa = 4.48TPDD282 pKa = 3.64AQPSTSAFTPAPGPPGYY299 pKa = 7.79PTAFTPAAGPPSSPSSFTPADD320 pKa = 4.13GPPSASPTVALNPNNVDD337 pKa = 3.12NAAPQATQFQPVSSPPVVQAPASTFTPAAAPSTQDD372 pKa = 3.14VASPATTQQIQPTPSSLVVQTAAPPVNSAPSFFTSASGVVTPTILATSSPVAADD426 pKa = 3.74TPTPAATSAPSSTVVDD442 pKa = 4.71TPATSSTVPLRR453 pKa = 11.84IYY455 pKa = 10.17VAAASPSSSSVDD467 pKa = 3.18AASSTTTPLTLVSLAPTAVSAQSAA491 pKa = 3.6
Molecular weight: 48.31 kDa
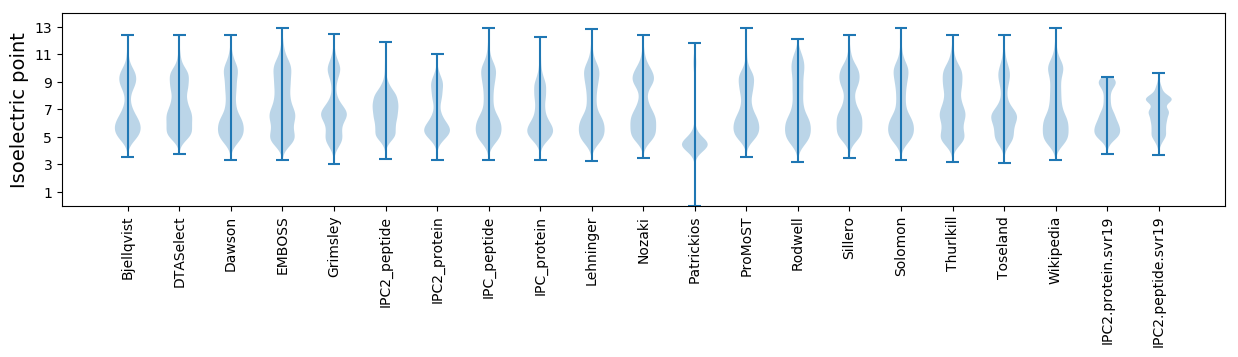
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0T047|G0T047_RHOT2 Serine/arginine repetitive matrix 1 OS=Rhodosporidium toruloides (strain ATCC 204091 / IIP 30 / MTCC 1151) OX=1001064 GN=RTG_02323 PE=4 SV=1
MM1 pKa = 7.65APLALQEE8 pKa = 4.07AHH10 pKa = 7.01KK11 pKa = 10.52SLKK14 pKa = 9.1TALTRR19 pKa = 11.84AHH21 pKa = 6.9GPACPFYY28 pKa = 10.69HH29 pKa = 7.17RR30 pKa = 11.84PQDD33 pKa = 3.51IPVAASLLVGLSALASPRR51 pKa = 11.84VPFRR55 pKa = 11.84TGSSGANRR63 pKa = 11.84LLVVLRR69 pKa = 11.84RR70 pKa = 11.84SAPSAQARR78 pKa = 11.84VQARR82 pKa = 11.84TFATSVRR89 pKa = 11.84TPSLAQGPRR98 pKa = 11.84GFEE101 pKa = 3.97AGLTGALKK109 pKa = 10.68GATGTSGVGRR119 pKa = 11.84AFQFLAGEE127 pKa = 4.32TGRR130 pKa = 11.84GILRR134 pKa = 11.84HH135 pKa = 5.22AKK137 pKa = 10.23ARR139 pKa = 11.84VEE141 pKa = 4.16SAEE144 pKa = 3.78RR145 pKa = 11.84LAANEE150 pKa = 3.98KK151 pKa = 8.76STRR154 pKa = 11.84TPWFWRR160 pKa = 11.84TTLPRR165 pKa = 11.84TGSFASRR172 pKa = 11.84RR173 pKa = 11.84DD174 pKa = 3.4QWTHH178 pKa = 4.93TLHH181 pKa = 7.21ASTSLKK187 pKa = 10.53CDD189 pKa = 4.1FAPSSTSLDD198 pKa = 3.23PSHH201 pKa = 7.22FNDD204 pKa = 3.94SSDD207 pKa = 3.11SRR209 pKa = 11.84SRR211 pKa = 11.84SRR213 pKa = 11.84SHH215 pKa = 6.21HH216 pKa = 6.07HH217 pKa = 3.58QHH219 pKa = 5.6QRR221 pKa = 11.84QSHH224 pKa = 5.95AEE226 pKa = 4.0SEE228 pKa = 4.38FDD230 pKa = 4.43SAHH233 pKa = 5.69QSCEE237 pKa = 4.05GQGGGQAAPVGGCRR251 pKa = 11.84SKK253 pKa = 11.19GPGWGWYY260 pKa = 9.02LALNEE265 pKa = 4.07RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84DD270 pKa = 3.57PQKK273 pKa = 9.9RR274 pKa = 11.84AHH276 pKa = 6.09FRR278 pKa = 11.84QVYY281 pKa = 10.02RR282 pKa = 11.84SMNPDD287 pKa = 3.11LKK289 pKa = 11.11DD290 pKa = 3.11PGMRR294 pKa = 11.84LCGQWEE300 pKa = 3.92AMRR303 pKa = 11.84WWEE306 pKa = 4.52RR307 pKa = 11.84RR308 pKa = 11.84LKK310 pKa = 10.32MEE312 pKa = 4.89GRR314 pKa = 11.84WKK316 pKa = 10.73RR317 pKa = 11.84NGGKK321 pKa = 9.2WSEE324 pKa = 4.83CYY326 pKa = 10.55EE327 pKa = 4.03EE328 pKa = 4.44MIKK331 pKa = 10.53EE332 pKa = 3.84RR333 pKa = 11.84DD334 pKa = 3.39VKK336 pKa = 10.68LRR338 pKa = 11.84RR339 pKa = 11.84LRR341 pKa = 11.84ALMVRR346 pKa = 11.84LRR348 pKa = 11.84AGAPNHH354 pKa = 5.57RR355 pKa = 11.84LRR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84MVDD362 pKa = 2.69SATGGRR368 pKa = 11.84YY369 pKa = 9.38GPSGSGPFNYY379 pKa = 9.9EE380 pKa = 3.49AFRR383 pKa = 11.84RR384 pKa = 11.84DD385 pKa = 3.25YY386 pKa = 9.9WRR388 pKa = 11.84SRR390 pKa = 11.84LAPHH394 pKa = 6.55RR395 pKa = 11.84QSGHH399 pKa = 4.74ARR401 pKa = 11.84RR402 pKa = 11.84SGAKK406 pKa = 9.33RR407 pKa = 11.84SASGFQWLALEE418 pKa = 4.1QRR420 pKa = 11.84RR421 pKa = 11.84RR422 pKa = 11.84RR423 pKa = 11.84AKK425 pKa = 10.34VGWAFFTWRR434 pKa = 11.84ACEE437 pKa = 4.19GPADD441 pKa = 3.45ARR443 pKa = 11.84KK444 pKa = 9.06WIRR447 pKa = 11.84RR448 pKa = 11.84DD449 pKa = 3.2GLVQKK454 pKa = 10.59SILGRR459 pKa = 11.84PQLLSAAPLARR470 pKa = 11.84PPSTILFATSAPRR483 pKa = 11.84LASATHH489 pKa = 5.27SAVRR493 pKa = 11.84AFSTSASRR501 pKa = 11.84QIPLPPPSLPLNILFAAPLCPTEE524 pKa = 4.27CALPLLLPLVSILKK538 pKa = 10.52SSAALNVLATITRR551 pKa = 11.84ISLTLLPLSFRR562 pKa = 11.84GKK564 pKa = 9.33ILHH567 pKa = 5.95SLRR570 pKa = 11.84EE571 pKa = 3.98RR572 pKa = 11.84YY573 pKa = 9.62VRR575 pKa = 11.84DD576 pKa = 3.58PTSLASSILGRR587 pKa = 11.84MVLNSGNAAQLAQPSSGFWRR607 pKa = 11.84WNALVGLPLLLATPLVLLALVALASLEE634 pKa = 4.04RR635 pKa = 11.84TPITGRR641 pKa = 11.84WRR643 pKa = 11.84VVMLSPAEE651 pKa = 3.93EE652 pKa = 4.5AEE654 pKa = 4.34LVDD657 pKa = 5.59SILSSADD664 pKa = 2.78KK665 pKa = 11.1SPFVTSPPEE674 pKa = 3.72GTTRR678 pKa = 11.84DD679 pKa = 3.13WVTILRR685 pKa = 11.84KK686 pKa = 9.91VLEE689 pKa = 4.67LPDD692 pKa = 4.31EE693 pKa = 4.81GVSSATGRR701 pKa = 11.84RR702 pKa = 11.84RR703 pKa = 11.84LLGGEE708 pKa = 4.26VLDD711 pKa = 3.89QRR713 pKa = 11.84DD714 pKa = 3.04WRR716 pKa = 11.84VRR718 pKa = 11.84WTQAVLEE725 pKa = 4.36ALEE728 pKa = 4.54KK729 pKa = 10.46GAPAALTASASSSSPSVMPPPPTAFPLEE757 pKa = 4.27PRR759 pKa = 11.84PEE761 pKa = 4.1ALGQGAAGWADD772 pKa = 3.19EE773 pKa = 4.85LVFSKK778 pKa = 10.57PLEE781 pKa = 4.15ARR783 pKa = 11.84HH784 pKa = 6.06ARR786 pKa = 11.84TEE788 pKa = 3.92HH789 pKa = 7.26DD790 pKa = 3.48EE791 pKa = 4.5GGVPLRR797 pKa = 11.84LEE799 pKa = 4.05YY800 pKa = 10.89DD801 pKa = 3.65LLVVDD806 pKa = 5.49RR807 pKa = 11.84KK808 pKa = 10.41DD809 pKa = 3.42ANAFSFGFGPDD820 pKa = 3.35EE821 pKa = 4.42VPSGRR826 pKa = 11.84AAKK829 pKa = 9.48PRR831 pKa = 11.84RR832 pKa = 11.84GVIVVYY838 pKa = 9.88TGFIDD843 pKa = 4.8EE844 pKa = 4.22ILGRR848 pKa = 11.84SGTDD852 pKa = 3.12VPLLSQPTPPSAKK865 pKa = 9.59RR866 pKa = 11.84SLLGFSRR873 pKa = 11.84STSPSTQPAPRR884 pKa = 11.84DD885 pKa = 3.66PIAANLVPPILPTDD899 pKa = 3.79AQTKK903 pKa = 9.68ALAVLLSHH911 pKa = 6.84EE912 pKa = 4.49LAHH915 pKa = 6.83LALSHH920 pKa = 5.68TLEE923 pKa = 5.05SYY925 pKa = 11.37ASTNLLVPHH934 pKa = 7.2LARR937 pKa = 11.84LTTDD941 pKa = 3.11VLRR944 pKa = 11.84TILYY948 pKa = 8.28PVTAILGPFINDD960 pKa = 3.47ALGGALNEE968 pKa = 4.37GARR971 pKa = 11.84GGLGVLGQAVNSCEE985 pKa = 4.07SRR987 pKa = 11.84KK988 pKa = 10.26LEE990 pKa = 4.16SEE992 pKa = 4.01ADD994 pKa = 3.79VVALRR999 pKa = 11.84MLATSGIDD1007 pKa = 3.18PHH1009 pKa = 6.9CALSFWEE1016 pKa = 5.03DD1017 pKa = 3.32RR1018 pKa = 11.84LSSPAHH1024 pKa = 6.33SPDD1027 pKa = 3.54SPTSSTHH1034 pKa = 5.7YY1035 pKa = 10.18PSLSQPNPLRR1045 pKa = 11.84PHH1047 pKa = 5.76STHH1050 pKa = 5.99EE1051 pKa = 4.06QDD1053 pKa = 5.2SKK1055 pKa = 11.94SLDD1058 pKa = 3.32ALLRR1062 pKa = 11.84SHH1064 pKa = 7.81PIDD1067 pKa = 3.38EE1068 pKa = 4.42EE1069 pKa = 3.88RR1070 pKa = 11.84VEE1072 pKa = 4.48RR1073 pKa = 11.84IRR1075 pKa = 11.84HH1076 pKa = 5.3EE1077 pKa = 3.97LRR1079 pKa = 11.84DD1080 pKa = 3.68WEE1082 pKa = 4.34KK1083 pKa = 9.55WWAEE1087 pKa = 4.1MKK1089 pKa = 10.29GHH1091 pKa = 5.82QAAAAA1096 pKa = 3.77
MM1 pKa = 7.65APLALQEE8 pKa = 4.07AHH10 pKa = 7.01KK11 pKa = 10.52SLKK14 pKa = 9.1TALTRR19 pKa = 11.84AHH21 pKa = 6.9GPACPFYY28 pKa = 10.69HH29 pKa = 7.17RR30 pKa = 11.84PQDD33 pKa = 3.51IPVAASLLVGLSALASPRR51 pKa = 11.84VPFRR55 pKa = 11.84TGSSGANRR63 pKa = 11.84LLVVLRR69 pKa = 11.84RR70 pKa = 11.84SAPSAQARR78 pKa = 11.84VQARR82 pKa = 11.84TFATSVRR89 pKa = 11.84TPSLAQGPRR98 pKa = 11.84GFEE101 pKa = 3.97AGLTGALKK109 pKa = 10.68GATGTSGVGRR119 pKa = 11.84AFQFLAGEE127 pKa = 4.32TGRR130 pKa = 11.84GILRR134 pKa = 11.84HH135 pKa = 5.22AKK137 pKa = 10.23ARR139 pKa = 11.84VEE141 pKa = 4.16SAEE144 pKa = 3.78RR145 pKa = 11.84LAANEE150 pKa = 3.98KK151 pKa = 8.76STRR154 pKa = 11.84TPWFWRR160 pKa = 11.84TTLPRR165 pKa = 11.84TGSFASRR172 pKa = 11.84RR173 pKa = 11.84DD174 pKa = 3.4QWTHH178 pKa = 4.93TLHH181 pKa = 7.21ASTSLKK187 pKa = 10.53CDD189 pKa = 4.1FAPSSTSLDD198 pKa = 3.23PSHH201 pKa = 7.22FNDD204 pKa = 3.94SSDD207 pKa = 3.11SRR209 pKa = 11.84SRR211 pKa = 11.84SRR213 pKa = 11.84SHH215 pKa = 6.21HH216 pKa = 6.07HH217 pKa = 3.58QHH219 pKa = 5.6QRR221 pKa = 11.84QSHH224 pKa = 5.95AEE226 pKa = 4.0SEE228 pKa = 4.38FDD230 pKa = 4.43SAHH233 pKa = 5.69QSCEE237 pKa = 4.05GQGGGQAAPVGGCRR251 pKa = 11.84SKK253 pKa = 11.19GPGWGWYY260 pKa = 9.02LALNEE265 pKa = 4.07RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84DD270 pKa = 3.57PQKK273 pKa = 9.9RR274 pKa = 11.84AHH276 pKa = 6.09FRR278 pKa = 11.84QVYY281 pKa = 10.02RR282 pKa = 11.84SMNPDD287 pKa = 3.11LKK289 pKa = 11.11DD290 pKa = 3.11PGMRR294 pKa = 11.84LCGQWEE300 pKa = 3.92AMRR303 pKa = 11.84WWEE306 pKa = 4.52RR307 pKa = 11.84RR308 pKa = 11.84LKK310 pKa = 10.32MEE312 pKa = 4.89GRR314 pKa = 11.84WKK316 pKa = 10.73RR317 pKa = 11.84NGGKK321 pKa = 9.2WSEE324 pKa = 4.83CYY326 pKa = 10.55EE327 pKa = 4.03EE328 pKa = 4.44MIKK331 pKa = 10.53EE332 pKa = 3.84RR333 pKa = 11.84DD334 pKa = 3.39VKK336 pKa = 10.68LRR338 pKa = 11.84RR339 pKa = 11.84LRR341 pKa = 11.84ALMVRR346 pKa = 11.84LRR348 pKa = 11.84AGAPNHH354 pKa = 5.57RR355 pKa = 11.84LRR357 pKa = 11.84RR358 pKa = 11.84RR359 pKa = 11.84MVDD362 pKa = 2.69SATGGRR368 pKa = 11.84YY369 pKa = 9.38GPSGSGPFNYY379 pKa = 9.9EE380 pKa = 3.49AFRR383 pKa = 11.84RR384 pKa = 11.84DD385 pKa = 3.25YY386 pKa = 9.9WRR388 pKa = 11.84SRR390 pKa = 11.84LAPHH394 pKa = 6.55RR395 pKa = 11.84QSGHH399 pKa = 4.74ARR401 pKa = 11.84RR402 pKa = 11.84SGAKK406 pKa = 9.33RR407 pKa = 11.84SASGFQWLALEE418 pKa = 4.1QRR420 pKa = 11.84RR421 pKa = 11.84RR422 pKa = 11.84RR423 pKa = 11.84AKK425 pKa = 10.34VGWAFFTWRR434 pKa = 11.84ACEE437 pKa = 4.19GPADD441 pKa = 3.45ARR443 pKa = 11.84KK444 pKa = 9.06WIRR447 pKa = 11.84RR448 pKa = 11.84DD449 pKa = 3.2GLVQKK454 pKa = 10.59SILGRR459 pKa = 11.84PQLLSAAPLARR470 pKa = 11.84PPSTILFATSAPRR483 pKa = 11.84LASATHH489 pKa = 5.27SAVRR493 pKa = 11.84AFSTSASRR501 pKa = 11.84QIPLPPPSLPLNILFAAPLCPTEE524 pKa = 4.27CALPLLLPLVSILKK538 pKa = 10.52SSAALNVLATITRR551 pKa = 11.84ISLTLLPLSFRR562 pKa = 11.84GKK564 pKa = 9.33ILHH567 pKa = 5.95SLRR570 pKa = 11.84EE571 pKa = 3.98RR572 pKa = 11.84YY573 pKa = 9.62VRR575 pKa = 11.84DD576 pKa = 3.58PTSLASSILGRR587 pKa = 11.84MVLNSGNAAQLAQPSSGFWRR607 pKa = 11.84WNALVGLPLLLATPLVLLALVALASLEE634 pKa = 4.04RR635 pKa = 11.84TPITGRR641 pKa = 11.84WRR643 pKa = 11.84VVMLSPAEE651 pKa = 3.93EE652 pKa = 4.5AEE654 pKa = 4.34LVDD657 pKa = 5.59SILSSADD664 pKa = 2.78KK665 pKa = 11.1SPFVTSPPEE674 pKa = 3.72GTTRR678 pKa = 11.84DD679 pKa = 3.13WVTILRR685 pKa = 11.84KK686 pKa = 9.91VLEE689 pKa = 4.67LPDD692 pKa = 4.31EE693 pKa = 4.81GVSSATGRR701 pKa = 11.84RR702 pKa = 11.84RR703 pKa = 11.84LLGGEE708 pKa = 4.26VLDD711 pKa = 3.89QRR713 pKa = 11.84DD714 pKa = 3.04WRR716 pKa = 11.84VRR718 pKa = 11.84WTQAVLEE725 pKa = 4.36ALEE728 pKa = 4.54KK729 pKa = 10.46GAPAALTASASSSSPSVMPPPPTAFPLEE757 pKa = 4.27PRR759 pKa = 11.84PEE761 pKa = 4.1ALGQGAAGWADD772 pKa = 3.19EE773 pKa = 4.85LVFSKK778 pKa = 10.57PLEE781 pKa = 4.15ARR783 pKa = 11.84HH784 pKa = 6.06ARR786 pKa = 11.84TEE788 pKa = 3.92HH789 pKa = 7.26DD790 pKa = 3.48EE791 pKa = 4.5GGVPLRR797 pKa = 11.84LEE799 pKa = 4.05YY800 pKa = 10.89DD801 pKa = 3.65LLVVDD806 pKa = 5.49RR807 pKa = 11.84KK808 pKa = 10.41DD809 pKa = 3.42ANAFSFGFGPDD820 pKa = 3.35EE821 pKa = 4.42VPSGRR826 pKa = 11.84AAKK829 pKa = 9.48PRR831 pKa = 11.84RR832 pKa = 11.84GVIVVYY838 pKa = 9.88TGFIDD843 pKa = 4.8EE844 pKa = 4.22ILGRR848 pKa = 11.84SGTDD852 pKa = 3.12VPLLSQPTPPSAKK865 pKa = 9.59RR866 pKa = 11.84SLLGFSRR873 pKa = 11.84STSPSTQPAPRR884 pKa = 11.84DD885 pKa = 3.66PIAANLVPPILPTDD899 pKa = 3.79AQTKK903 pKa = 9.68ALAVLLSHH911 pKa = 6.84EE912 pKa = 4.49LAHH915 pKa = 6.83LALSHH920 pKa = 5.68TLEE923 pKa = 5.05SYY925 pKa = 11.37ASTNLLVPHH934 pKa = 7.2LARR937 pKa = 11.84LTTDD941 pKa = 3.11VLRR944 pKa = 11.84TILYY948 pKa = 8.28PVTAILGPFINDD960 pKa = 3.47ALGGALNEE968 pKa = 4.37GARR971 pKa = 11.84GGLGVLGQAVNSCEE985 pKa = 4.07SRR987 pKa = 11.84KK988 pKa = 10.26LEE990 pKa = 4.16SEE992 pKa = 4.01ADD994 pKa = 3.79VVALRR999 pKa = 11.84MLATSGIDD1007 pKa = 3.18PHH1009 pKa = 6.9CALSFWEE1016 pKa = 5.03DD1017 pKa = 3.32RR1018 pKa = 11.84LSSPAHH1024 pKa = 6.33SPDD1027 pKa = 3.54SPTSSTHH1034 pKa = 5.7YY1035 pKa = 10.18PSLSQPNPLRR1045 pKa = 11.84PHH1047 pKa = 5.76STHH1050 pKa = 5.99EE1051 pKa = 4.06QDD1053 pKa = 5.2SKK1055 pKa = 11.94SLDD1058 pKa = 3.32ALLRR1062 pKa = 11.84SHH1064 pKa = 7.81PIDD1067 pKa = 3.38EE1068 pKa = 4.42EE1069 pKa = 3.88RR1070 pKa = 11.84VEE1072 pKa = 4.48RR1073 pKa = 11.84IRR1075 pKa = 11.84HH1076 pKa = 5.3EE1077 pKa = 3.97LRR1079 pKa = 11.84DD1080 pKa = 3.68WEE1082 pKa = 4.34KK1083 pKa = 9.55WWAEE1087 pKa = 4.1MKK1089 pKa = 10.29GHH1091 pKa = 5.82QAAAAA1096 pKa = 3.77
Molecular weight: 120.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1800369 |
29 |
4364 |
639.3 |
69.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.832 ± 0.043 | 1.008 ± 0.016 |
5.488 ± 0.027 | 6.452 ± 0.039 |
3.29 ± 0.024 | 6.912 ± 0.04 |
2.217 ± 0.015 | 3.348 ± 0.023 |
4.231 ± 0.033 | 9.537 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.575 ± 0.012 | 2.425 ± 0.017 |
7.394 ± 0.052 | 3.566 ± 0.026 |
7.026 ± 0.033 | 9.429 ± 0.052 |
5.777 ± 0.024 | 6.111 ± 0.031 |
1.265 ± 0.014 | 2.118 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
