
Sphingopyxis sp. Root1497
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingopyxis; unclassified Sphingopyxis
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
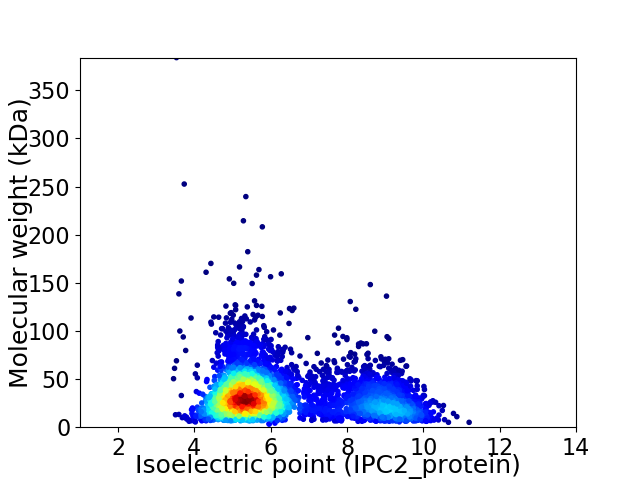
Virtual 2D-PAGE plot for 4384 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7YLG1|A0A0Q7YLG1_9SPHN Cytochrome OS=Sphingopyxis sp. Root1497 OX=1736474 GN=ASD67_12080 PE=3 SV=1
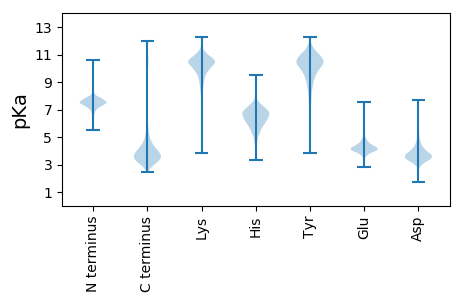
MM1 pKa = 7.05VLSTRR6 pKa = 11.84LQTVGEE12 pKa = 4.28SGNVFIDD19 pKa = 3.36TLVYY23 pKa = 10.4GVAFRR28 pKa = 11.84PATTVIYY35 pKa = 10.32ALQGNPGDD43 pKa = 3.66TGLNGGALWAANGAAGAYY61 pKa = 8.9AAALASWAAVANIRR75 pKa = 11.84FQAATTAYY83 pKa = 10.18DD84 pKa = 3.74GTGSRR89 pKa = 11.84ASYY92 pKa = 11.09DD93 pKa = 3.34FVGQLSTSLPSDD105 pKa = 3.29VLGQHH110 pKa = 6.31TLPTTGDD117 pKa = 3.15MVGQFNTTISYY128 pKa = 7.45FTTASNQAGGLSFLTFVHH146 pKa = 6.29EE147 pKa = 4.48LGHH150 pKa = 6.94GIGLLHH156 pKa = 6.58PHH158 pKa = 6.94ADD160 pKa = 3.72EE161 pKa = 5.56PGDD164 pKa = 3.99DD165 pKa = 4.05SFPGLGDD172 pKa = 3.72AFSTGPFGLNQGIFTTMTYY191 pKa = 10.8NDD193 pKa = 4.11GYY195 pKa = 10.03EE196 pKa = 4.15DD197 pKa = 3.54VGRR200 pKa = 11.84SPSVNYY206 pKa = 8.8GWQMGPGAFDD216 pKa = 3.15IAAVQFLYY224 pKa = 10.7GANNSTGAGNTTYY237 pKa = 11.03TMPTLNQAGTGWMTIWDD254 pKa = 3.85VGGTDD259 pKa = 3.78TISAQGSQLDD269 pKa = 4.03AIIDD273 pKa = 3.65LRR275 pKa = 11.84AATLRR280 pKa = 11.84NEE282 pKa = 4.07EE283 pKa = 4.14GGGGFVSRR291 pKa = 11.84NGGVLGGFTIANGAVIEE308 pKa = 4.08NAIGGNGDD316 pKa = 3.88DD317 pKa = 4.64LLVGNAANNSLNGGAGWDD335 pKa = 3.86TVSYY339 pKa = 11.36AHH341 pKa = 6.29MTSAVTVNLATGIATGEE358 pKa = 4.3GNDD361 pKa = 3.57TLSAIEE367 pKa = 3.95RR368 pKa = 11.84AIGGSGDD375 pKa = 3.88DD376 pKa = 3.93VMTAFIGASIVNGSQDD392 pKa = 3.8VIKK395 pKa = 10.8SAALRR400 pKa = 11.84NGEE403 pKa = 4.06IASALSLDD411 pKa = 4.15GRR413 pKa = 11.84FSPHH417 pKa = 6.54SSDD420 pKa = 3.94VSIQAVGAGVNSVTVHH436 pKa = 6.42GEE438 pKa = 3.67GGLQRR443 pKa = 11.84DD444 pKa = 4.53VYY446 pKa = 11.3SFTLTAGAQITIDD459 pKa = 3.22IDD461 pKa = 3.23NSFRR465 pKa = 11.84MDD467 pKa = 4.09SIIALFGPGLYY478 pKa = 10.47GPSSLMTTNDD488 pKa = 4.07DD489 pKa = 3.93SPNDD493 pKa = 3.94LDD495 pKa = 3.89VGSANSLDD503 pKa = 3.68SFLTFNATTAGTYY516 pKa = 10.02YY517 pKa = 11.35VMVSTYY523 pKa = 10.19SAEE526 pKa = 4.14SDD528 pKa = 3.33GGGPIALGSSYY539 pKa = 10.57TLNISAANTVAAAGTILLGSTLDD562 pKa = 3.74GGAGNDD568 pKa = 4.0TLNGGAGIDD577 pKa = 3.73TLIGGIGNDD586 pKa = 3.55MLNGGGGADD595 pKa = 3.86LMYY598 pKa = 10.97GGTGNDD604 pKa = 3.26NYY606 pKa = 10.84VIDD609 pKa = 4.36AQGDD613 pKa = 3.87LTFEE617 pKa = 4.33GLNGGTDD624 pKa = 3.26TVTASVGHH632 pKa = 5.55YY633 pKa = 10.35LYY635 pKa = 11.29ANLEE639 pKa = 4.1NLTLATGAGDD649 pKa = 3.14IFGVGNEE656 pKa = 4.68LANTLQGNAGANLLLGGLGDD676 pKa = 3.86DD677 pKa = 4.87VIRR680 pKa = 11.84GGAGVDD686 pKa = 3.61SLFGEE691 pKa = 4.58AGVDD695 pKa = 3.77QLFGDD700 pKa = 3.94AGIDD704 pKa = 3.67YY705 pKa = 10.82LVGGLGNDD713 pKa = 3.55ILDD716 pKa = 4.2GGADD720 pKa = 3.46ADD722 pKa = 3.93ALYY725 pKa = 11.17GEE727 pKa = 5.66DD728 pKa = 5.55GDD730 pKa = 4.24DD731 pKa = 3.72TLIGGAGFFTDD742 pKa = 3.79ILVGGAGNDD751 pKa = 3.31ILRR754 pKa = 11.84GASGLGDD761 pKa = 4.2YY762 pKa = 11.48DD763 pKa = 5.77LMDD766 pKa = 4.59GGAGNDD772 pKa = 3.02IYY774 pKa = 11.66YY775 pKa = 10.51VDD777 pKa = 4.51TPDD780 pKa = 4.83DD781 pKa = 3.77LTFEE785 pKa = 4.39AVGGGADD792 pKa = 2.97TVYY795 pKa = 11.49ANINGAGYY803 pKa = 9.84YY804 pKa = 10.35LYY806 pKa = 11.15ANVEE810 pKa = 4.15NLVLEE815 pKa = 4.69GNTPFGVGNDD825 pKa = 3.41LANRR829 pKa = 11.84LTGNAIGNYY838 pKa = 9.89LLGGRR843 pKa = 11.84GNDD846 pKa = 3.47TLNGKK851 pKa = 9.26GGNDD855 pKa = 3.28VLFGEE860 pKa = 5.0GGVDD864 pKa = 2.93TFVFEE869 pKa = 4.81RR870 pKa = 11.84GTGGDD875 pKa = 3.59VIGDD879 pKa = 4.09FARR882 pKa = 11.84GTDD885 pKa = 4.06KK886 pKa = 10.93IDD888 pKa = 3.14VSAFGFASFAALQAGFTQVGANGAINLGNGDD919 pKa = 4.95FIVLHH924 pKa = 6.05NVTMSQLTQGDD935 pKa = 4.93FILSATAGKK944 pKa = 9.44PDD946 pKa = 4.32LFGDD950 pKa = 3.96GAKK953 pKa = 10.24AVEE956 pKa = 4.47DD957 pKa = 3.79AGFAPDD963 pKa = 4.11AGTPVLDD970 pKa = 3.38WHH972 pKa = 7.14PDD974 pKa = 2.57HH975 pKa = 7.41WMAQYY980 pKa = 8.26HH981 pKa = 5.14TAYY984 pKa = 10.69QLGFVV989 pKa = 4.32
MM1 pKa = 7.05VLSTRR6 pKa = 11.84LQTVGEE12 pKa = 4.28SGNVFIDD19 pKa = 3.36TLVYY23 pKa = 10.4GVAFRR28 pKa = 11.84PATTVIYY35 pKa = 10.32ALQGNPGDD43 pKa = 3.66TGLNGGALWAANGAAGAYY61 pKa = 8.9AAALASWAAVANIRR75 pKa = 11.84FQAATTAYY83 pKa = 10.18DD84 pKa = 3.74GTGSRR89 pKa = 11.84ASYY92 pKa = 11.09DD93 pKa = 3.34FVGQLSTSLPSDD105 pKa = 3.29VLGQHH110 pKa = 6.31TLPTTGDD117 pKa = 3.15MVGQFNTTISYY128 pKa = 7.45FTTASNQAGGLSFLTFVHH146 pKa = 6.29EE147 pKa = 4.48LGHH150 pKa = 6.94GIGLLHH156 pKa = 6.58PHH158 pKa = 6.94ADD160 pKa = 3.72EE161 pKa = 5.56PGDD164 pKa = 3.99DD165 pKa = 4.05SFPGLGDD172 pKa = 3.72AFSTGPFGLNQGIFTTMTYY191 pKa = 10.8NDD193 pKa = 4.11GYY195 pKa = 10.03EE196 pKa = 4.15DD197 pKa = 3.54VGRR200 pKa = 11.84SPSVNYY206 pKa = 8.8GWQMGPGAFDD216 pKa = 3.15IAAVQFLYY224 pKa = 10.7GANNSTGAGNTTYY237 pKa = 11.03TMPTLNQAGTGWMTIWDD254 pKa = 3.85VGGTDD259 pKa = 3.78TISAQGSQLDD269 pKa = 4.03AIIDD273 pKa = 3.65LRR275 pKa = 11.84AATLRR280 pKa = 11.84NEE282 pKa = 4.07EE283 pKa = 4.14GGGGFVSRR291 pKa = 11.84NGGVLGGFTIANGAVIEE308 pKa = 4.08NAIGGNGDD316 pKa = 3.88DD317 pKa = 4.64LLVGNAANNSLNGGAGWDD335 pKa = 3.86TVSYY339 pKa = 11.36AHH341 pKa = 6.29MTSAVTVNLATGIATGEE358 pKa = 4.3GNDD361 pKa = 3.57TLSAIEE367 pKa = 3.95RR368 pKa = 11.84AIGGSGDD375 pKa = 3.88DD376 pKa = 3.93VMTAFIGASIVNGSQDD392 pKa = 3.8VIKK395 pKa = 10.8SAALRR400 pKa = 11.84NGEE403 pKa = 4.06IASALSLDD411 pKa = 4.15GRR413 pKa = 11.84FSPHH417 pKa = 6.54SSDD420 pKa = 3.94VSIQAVGAGVNSVTVHH436 pKa = 6.42GEE438 pKa = 3.67GGLQRR443 pKa = 11.84DD444 pKa = 4.53VYY446 pKa = 11.3SFTLTAGAQITIDD459 pKa = 3.22IDD461 pKa = 3.23NSFRR465 pKa = 11.84MDD467 pKa = 4.09SIIALFGPGLYY478 pKa = 10.47GPSSLMTTNDD488 pKa = 4.07DD489 pKa = 3.93SPNDD493 pKa = 3.94LDD495 pKa = 3.89VGSANSLDD503 pKa = 3.68SFLTFNATTAGTYY516 pKa = 10.02YY517 pKa = 11.35VMVSTYY523 pKa = 10.19SAEE526 pKa = 4.14SDD528 pKa = 3.33GGGPIALGSSYY539 pKa = 10.57TLNISAANTVAAAGTILLGSTLDD562 pKa = 3.74GGAGNDD568 pKa = 4.0TLNGGAGIDD577 pKa = 3.73TLIGGIGNDD586 pKa = 3.55MLNGGGGADD595 pKa = 3.86LMYY598 pKa = 10.97GGTGNDD604 pKa = 3.26NYY606 pKa = 10.84VIDD609 pKa = 4.36AQGDD613 pKa = 3.87LTFEE617 pKa = 4.33GLNGGTDD624 pKa = 3.26TVTASVGHH632 pKa = 5.55YY633 pKa = 10.35LYY635 pKa = 11.29ANLEE639 pKa = 4.1NLTLATGAGDD649 pKa = 3.14IFGVGNEE656 pKa = 4.68LANTLQGNAGANLLLGGLGDD676 pKa = 3.86DD677 pKa = 4.87VIRR680 pKa = 11.84GGAGVDD686 pKa = 3.61SLFGEE691 pKa = 4.58AGVDD695 pKa = 3.77QLFGDD700 pKa = 3.94AGIDD704 pKa = 3.67YY705 pKa = 10.82LVGGLGNDD713 pKa = 3.55ILDD716 pKa = 4.2GGADD720 pKa = 3.46ADD722 pKa = 3.93ALYY725 pKa = 11.17GEE727 pKa = 5.66DD728 pKa = 5.55GDD730 pKa = 4.24DD731 pKa = 3.72TLIGGAGFFTDD742 pKa = 3.79ILVGGAGNDD751 pKa = 3.31ILRR754 pKa = 11.84GASGLGDD761 pKa = 4.2YY762 pKa = 11.48DD763 pKa = 5.77LMDD766 pKa = 4.59GGAGNDD772 pKa = 3.02IYY774 pKa = 11.66YY775 pKa = 10.51VDD777 pKa = 4.51TPDD780 pKa = 4.83DD781 pKa = 3.77LTFEE785 pKa = 4.39AVGGGADD792 pKa = 2.97TVYY795 pKa = 11.49ANINGAGYY803 pKa = 9.84YY804 pKa = 10.35LYY806 pKa = 11.15ANVEE810 pKa = 4.15NLVLEE815 pKa = 4.69GNTPFGVGNDD825 pKa = 3.41LANRR829 pKa = 11.84LTGNAIGNYY838 pKa = 9.89LLGGRR843 pKa = 11.84GNDD846 pKa = 3.47TLNGKK851 pKa = 9.26GGNDD855 pKa = 3.28VLFGEE860 pKa = 5.0GGVDD864 pKa = 2.93TFVFEE869 pKa = 4.81RR870 pKa = 11.84GTGGDD875 pKa = 3.59VIGDD879 pKa = 4.09FARR882 pKa = 11.84GTDD885 pKa = 4.06KK886 pKa = 10.93IDD888 pKa = 3.14VSAFGFASFAALQAGFTQVGANGAINLGNGDD919 pKa = 4.95FIVLHH924 pKa = 6.05NVTMSQLTQGDD935 pKa = 4.93FILSATAGKK944 pKa = 9.44PDD946 pKa = 4.32LFGDD950 pKa = 3.96GAKK953 pKa = 10.24AVEE956 pKa = 4.47DD957 pKa = 3.79AGFAPDD963 pKa = 4.11AGTPVLDD970 pKa = 3.38WHH972 pKa = 7.14PDD974 pKa = 2.57HH975 pKa = 7.41WMAQYY980 pKa = 8.26HH981 pKa = 5.14TAYY984 pKa = 10.69QLGFVV989 pKa = 4.32
Molecular weight: 99.92 kDa
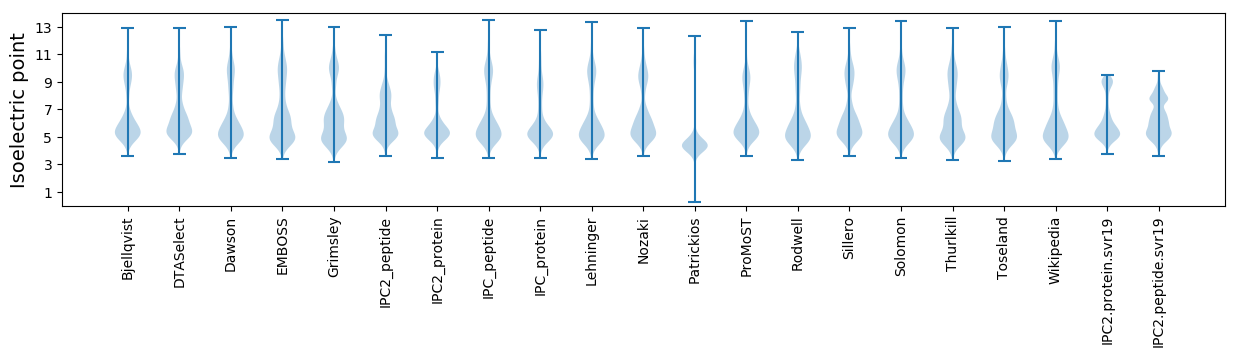
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7YY40|A0A0Q7YY40_9SPHN Pimeloyl-CoA dehydrogenase large subunit OS=Sphingopyxis sp. Root1497 OX=1736474 GN=ASD67_22015 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.43GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 8.83ILANRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.26KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.43GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 8.83ILANRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.26KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1426498 |
29 |
4039 |
325.4 |
35.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.824 ± 0.064 | 0.757 ± 0.01 |
6.305 ± 0.027 | 5.121 ± 0.03 |
3.614 ± 0.027 | 9.205 ± 0.054 |
1.949 ± 0.019 | 5.106 ± 0.023 |
3.014 ± 0.033 | 9.679 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.02 | 2.557 ± 0.029 |
5.326 ± 0.029 | 2.906 ± 0.021 |
7.004 ± 0.035 | 5.121 ± 0.028 |
5.273 ± 0.032 | 7.051 ± 0.028 |
1.497 ± 0.017 | 2.279 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
