
Runella sp. YX9
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Runella
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
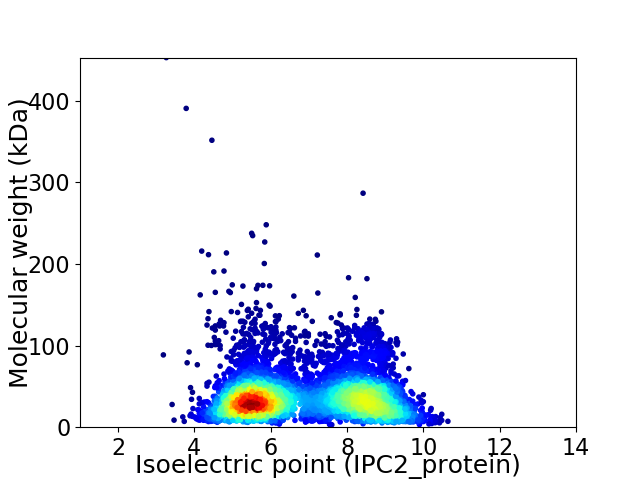
Virtual 2D-PAGE plot for 5856 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369IJQ5|A0A369IJQ5_9BACT VCBS repeat-containing protein OS=Runella sp. YX9 OX=2282308 GN=DVG78_00665 PE=4 SV=1
AA1 pKa = 7.5TFLSPVWTVGTANGTVVATPTSVQPITTTTYY32 pKa = 11.26VLVAQNAEE40 pKa = 4.28GCQDD44 pKa = 3.35TSQVVVTVNAKK55 pKa = 9.95PNAGRR60 pKa = 11.84DD61 pKa = 3.51TTLACVDD68 pKa = 3.84ALSNTLATSYY78 pKa = 9.8TLVPIPPGGTWSQIGTTPTTATVTGNNVTGMTMAGTYY115 pKa = 8.79QFIYY119 pKa = 9.8TLNDD123 pKa = 3.11CSDD126 pKa = 3.94TVSVTVSPCQGCVQPNAGPDD146 pKa = 3.39QSICSPTTTATLTAITLGGTWTAQLGNPAPATITNLGAVSGMTIDD191 pKa = 3.29GTYY194 pKa = 10.52RR195 pKa = 11.84FIYY198 pKa = 10.19SVTSGGQTCTDD209 pKa = 3.38TVSVIRR215 pKa = 11.84NPKK218 pKa = 9.21PEE220 pKa = 4.83AGPDD224 pKa = 3.43QSICSPTTTATLTAITLGGTWSAQSGNPVAATITNAGTVTGMTANGTYY272 pKa = 10.83NFIYY276 pKa = 9.26MINGCTDD283 pKa = 3.15TVSVVRR289 pKa = 11.84TAKK292 pKa = 10.09PDD294 pKa = 3.32AGADD298 pKa = 3.53QTLTCPPSGISPTTANVTGAPSGGVWSTLASNPAAATVTNAGAVSGMTVAGTYY351 pKa = 8.82QFIYY355 pKa = 9.8TLNDD359 pKa = 3.09CSDD362 pKa = 3.63TVQIQVPTCIAPVFDD377 pKa = 4.39LALSKK382 pKa = 9.72TLAVGQSASVVAGSTVTFTITVFNQGNVDD411 pKa = 3.27ATNIQVTDD419 pKa = 4.2YY420 pKa = 10.84IPAGLTLNDD429 pKa = 4.37ANWSVVGSTATLNTVIASLAAGASTTRR456 pKa = 11.84DD457 pKa = 2.71ITFVVSTGVTGSITNLAEE475 pKa = 3.99ISSATGGVDD484 pKa = 3.16VDD486 pKa = 4.7SSPDD490 pKa = 3.51SNPANDD496 pKa = 4.15GTPKK500 pKa = 10.65NDD502 pKa = 3.55VINEE506 pKa = 3.96NGLTGGDD513 pKa = 4.08EE514 pKa = 4.62DD515 pKa = 4.7DD516 pKa = 5.1HH517 pKa = 8.83DD518 pKa = 4.81PEE520 pKa = 5.54VITVTPAPVFDD531 pKa = 3.84LALRR535 pKa = 11.84KK536 pKa = 7.82TLAVGQSASVVAGSTVTFTITVFNQGNVDD565 pKa = 3.27ATNIQVTDD573 pKa = 4.2YY574 pKa = 10.84IPAGLTLNDD583 pKa = 4.37ANWSVVGSTATLNTVIASLAAGASTTRR610 pKa = 11.84DD611 pKa = 2.71ITFVVSTGVTGSLSNLAEE629 pKa = 4.14ISSATGGVDD638 pKa = 3.16VDD640 pKa = 4.7SSPDD644 pKa = 3.53SNPADD649 pKa = 4.52DD650 pKa = 4.6GTPKK654 pKa = 10.64NDD656 pKa = 3.58VINEE660 pKa = 4.08SGLTGGDD667 pKa = 3.7EE668 pKa = 4.64DD669 pKa = 4.7DD670 pKa = 5.1HH671 pKa = 8.83DD672 pKa = 4.81PEE674 pKa = 5.47VITVTSAQVDD684 pKa = 4.11LSLKK688 pKa = 10.72KK689 pKa = 10.1MISTKK694 pKa = 9.92IVQLGDD700 pKa = 3.2TLTYY704 pKa = 8.81TIKK707 pKa = 10.63VFNQSGTLATGVEE720 pKa = 4.59VSDD723 pKa = 4.1SLATSVQFIMGSFTTTRR740 pKa = 11.84GTATLSNNVIVWTIGSIGANGDD762 pKa = 4.03TVTLTYY768 pKa = 9.96KK769 pKa = 10.61VKK771 pKa = 10.56AISEE775 pKa = 4.28GVHH778 pKa = 6.23FNTAEE783 pKa = 3.66ISKK786 pKa = 9.29TNEE789 pKa = 3.71PDD791 pKa = 3.16VDD793 pKa = 3.95STPGNGVGNEE803 pKa = 4.01DD804 pKa = 5.45DD805 pKa = 4.66IDD807 pKa = 3.99RR808 pKa = 11.84QCFTVPLKK816 pKa = 11.04LCVGEE821 pKa = 4.39KK822 pKa = 10.04IEE824 pKa = 4.19VSVPSYY830 pKa = 10.54LVNVQWYY837 pKa = 9.18KK838 pKa = 11.3DD839 pKa = 3.32GGTIPVVSGNVVLFSEE855 pKa = 4.6AGSYY859 pKa = 9.09TFTASNQQCPATGCCPIIIEE879 pKa = 4.97PGDD882 pKa = 3.46NCCPVDD888 pKa = 3.79LCIPFTIKK896 pKa = 8.34QTKK899 pKa = 9.41KK900 pKa = 10.36AGKK903 pKa = 8.71PLL905 pKa = 3.66
AA1 pKa = 7.5TFLSPVWTVGTANGTVVATPTSVQPITTTTYY32 pKa = 11.26VLVAQNAEE40 pKa = 4.28GCQDD44 pKa = 3.35TSQVVVTVNAKK55 pKa = 9.95PNAGRR60 pKa = 11.84DD61 pKa = 3.51TTLACVDD68 pKa = 3.84ALSNTLATSYY78 pKa = 9.8TLVPIPPGGTWSQIGTTPTTATVTGNNVTGMTMAGTYY115 pKa = 8.79QFIYY119 pKa = 9.8TLNDD123 pKa = 3.11CSDD126 pKa = 3.94TVSVTVSPCQGCVQPNAGPDD146 pKa = 3.39QSICSPTTTATLTAITLGGTWTAQLGNPAPATITNLGAVSGMTIDD191 pKa = 3.29GTYY194 pKa = 10.52RR195 pKa = 11.84FIYY198 pKa = 10.19SVTSGGQTCTDD209 pKa = 3.38TVSVIRR215 pKa = 11.84NPKK218 pKa = 9.21PEE220 pKa = 4.83AGPDD224 pKa = 3.43QSICSPTTTATLTAITLGGTWSAQSGNPVAATITNAGTVTGMTANGTYY272 pKa = 10.83NFIYY276 pKa = 9.26MINGCTDD283 pKa = 3.15TVSVVRR289 pKa = 11.84TAKK292 pKa = 10.09PDD294 pKa = 3.32AGADD298 pKa = 3.53QTLTCPPSGISPTTANVTGAPSGGVWSTLASNPAAATVTNAGAVSGMTVAGTYY351 pKa = 8.82QFIYY355 pKa = 9.8TLNDD359 pKa = 3.09CSDD362 pKa = 3.63TVQIQVPTCIAPVFDD377 pKa = 4.39LALSKK382 pKa = 9.72TLAVGQSASVVAGSTVTFTITVFNQGNVDD411 pKa = 3.27ATNIQVTDD419 pKa = 4.2YY420 pKa = 10.84IPAGLTLNDD429 pKa = 4.37ANWSVVGSTATLNTVIASLAAGASTTRR456 pKa = 11.84DD457 pKa = 2.71ITFVVSTGVTGSITNLAEE475 pKa = 3.99ISSATGGVDD484 pKa = 3.16VDD486 pKa = 4.7SSPDD490 pKa = 3.51SNPANDD496 pKa = 4.15GTPKK500 pKa = 10.65NDD502 pKa = 3.55VINEE506 pKa = 3.96NGLTGGDD513 pKa = 4.08EE514 pKa = 4.62DD515 pKa = 4.7DD516 pKa = 5.1HH517 pKa = 8.83DD518 pKa = 4.81PEE520 pKa = 5.54VITVTPAPVFDD531 pKa = 3.84LALRR535 pKa = 11.84KK536 pKa = 7.82TLAVGQSASVVAGSTVTFTITVFNQGNVDD565 pKa = 3.27ATNIQVTDD573 pKa = 4.2YY574 pKa = 10.84IPAGLTLNDD583 pKa = 4.37ANWSVVGSTATLNTVIASLAAGASTTRR610 pKa = 11.84DD611 pKa = 2.71ITFVVSTGVTGSLSNLAEE629 pKa = 4.14ISSATGGVDD638 pKa = 3.16VDD640 pKa = 4.7SSPDD644 pKa = 3.53SNPADD649 pKa = 4.52DD650 pKa = 4.6GTPKK654 pKa = 10.64NDD656 pKa = 3.58VINEE660 pKa = 4.08SGLTGGDD667 pKa = 3.7EE668 pKa = 4.64DD669 pKa = 4.7DD670 pKa = 5.1HH671 pKa = 8.83DD672 pKa = 4.81PEE674 pKa = 5.47VITVTSAQVDD684 pKa = 4.11LSLKK688 pKa = 10.72KK689 pKa = 10.1MISTKK694 pKa = 9.92IVQLGDD700 pKa = 3.2TLTYY704 pKa = 8.81TIKK707 pKa = 10.63VFNQSGTLATGVEE720 pKa = 4.59VSDD723 pKa = 4.1SLATSVQFIMGSFTTTRR740 pKa = 11.84GTATLSNNVIVWTIGSIGANGDD762 pKa = 4.03TVTLTYY768 pKa = 9.96KK769 pKa = 10.61VKK771 pKa = 10.56AISEE775 pKa = 4.28GVHH778 pKa = 6.23FNTAEE783 pKa = 3.66ISKK786 pKa = 9.29TNEE789 pKa = 3.71PDD791 pKa = 3.16VDD793 pKa = 3.95STPGNGVGNEE803 pKa = 4.01DD804 pKa = 5.45DD805 pKa = 4.66IDD807 pKa = 3.99RR808 pKa = 11.84QCFTVPLKK816 pKa = 11.04LCVGEE821 pKa = 4.39KK822 pKa = 10.04IEE824 pKa = 4.19VSVPSYY830 pKa = 10.54LVNVQWYY837 pKa = 9.18KK838 pKa = 11.3DD839 pKa = 3.32GGTIPVVSGNVVLFSEE855 pKa = 4.6AGSYY859 pKa = 9.09TFTASNQQCPATGCCPIIIEE879 pKa = 4.97PGDD882 pKa = 3.46NCCPVDD888 pKa = 3.79LCIPFTIKK896 pKa = 8.34QTKK899 pKa = 9.41KK900 pKa = 10.36AGKK903 pKa = 8.71PLL905 pKa = 3.66
Molecular weight: 92.24 kDa
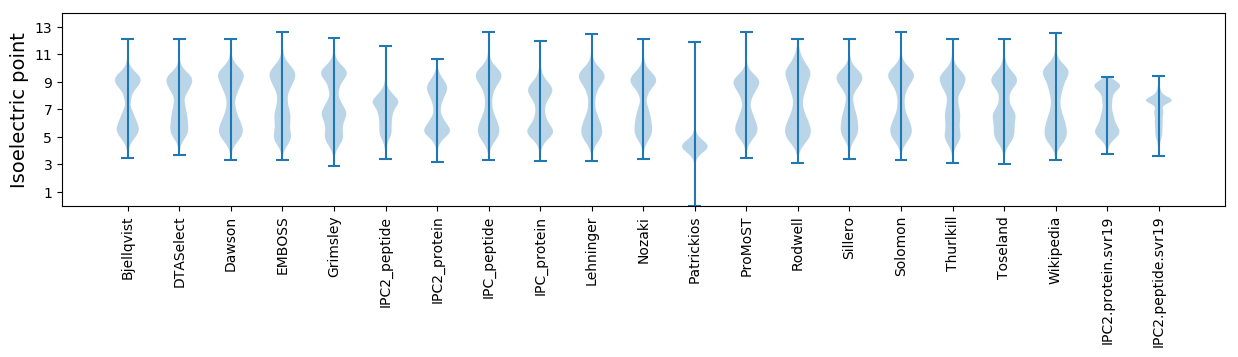
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369IIA9|A0A369IIA9_9BACT Uncharacterized protein OS=Runella sp. YX9 OX=2282308 GN=DVG78_06885 PE=4 SV=1
MM1 pKa = 6.98KK2 pKa = 10.28QVIAYY7 pKa = 9.0YY8 pKa = 10.02RR9 pKa = 11.84VSTQRR14 pKa = 11.84QGRR17 pKa = 11.84SGLGLEE23 pKa = 4.33AQQNAVLNYY32 pKa = 9.18CKK34 pKa = 10.53LSGYY38 pKa = 8.11EE39 pKa = 3.93VIRR42 pKa = 11.84EE43 pKa = 4.06VTEE46 pKa = 4.13VKK48 pKa = 9.24STRR51 pKa = 11.84KK52 pKa = 10.02DD53 pKa = 3.39RR54 pKa = 11.84IGLLQALDD62 pKa = 3.98LCRR65 pKa = 11.84QMKK68 pKa = 10.29AILMVARR75 pKa = 11.84LDD77 pKa = 3.59RR78 pKa = 11.84LGRR81 pKa = 11.84DD82 pKa = 3.5VEE84 pKa = 4.41QIAGIVKK91 pKa = 10.1SKK93 pKa = 11.26VEE95 pKa = 4.63IIVTDD100 pKa = 3.57NPHH103 pKa = 6.55ANRR106 pKa = 11.84FTIHH110 pKa = 6.35ILAAVAEE117 pKa = 4.41DD118 pKa = 3.18NRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84ISEE125 pKa = 4.23TTKK128 pKa = 10.84DD129 pKa = 3.37ALGAAKK135 pKa = 10.15RR136 pKa = 11.84RR137 pKa = 11.84GVILGKK143 pKa = 10.36NGTALSIANKK153 pKa = 9.73KK154 pKa = 9.32AAEE157 pKa = 4.42DD158 pKa = 3.85FAQQLSPTLKK168 pKa = 9.69RR169 pKa = 11.84LKK171 pKa = 9.34TRR173 pKa = 11.84GIISVRR179 pKa = 11.84AVCRR183 pKa = 11.84EE184 pKa = 3.84LNKK187 pKa = 10.44RR188 pKa = 11.84QIPTFRR194 pKa = 11.84GDD196 pKa = 4.0GRR198 pKa = 11.84WHH200 pKa = 6.95PSTVNVLMNRR210 pKa = 11.84LKK212 pKa = 9.68QTIISPP218 pKa = 4.03
MM1 pKa = 6.98KK2 pKa = 10.28QVIAYY7 pKa = 9.0YY8 pKa = 10.02RR9 pKa = 11.84VSTQRR14 pKa = 11.84QGRR17 pKa = 11.84SGLGLEE23 pKa = 4.33AQQNAVLNYY32 pKa = 9.18CKK34 pKa = 10.53LSGYY38 pKa = 8.11EE39 pKa = 3.93VIRR42 pKa = 11.84EE43 pKa = 4.06VTEE46 pKa = 4.13VKK48 pKa = 9.24STRR51 pKa = 11.84KK52 pKa = 10.02DD53 pKa = 3.39RR54 pKa = 11.84IGLLQALDD62 pKa = 3.98LCRR65 pKa = 11.84QMKK68 pKa = 10.29AILMVARR75 pKa = 11.84LDD77 pKa = 3.59RR78 pKa = 11.84LGRR81 pKa = 11.84DD82 pKa = 3.5VEE84 pKa = 4.41QIAGIVKK91 pKa = 10.1SKK93 pKa = 11.26VEE95 pKa = 4.63IIVTDD100 pKa = 3.57NPHH103 pKa = 6.55ANRR106 pKa = 11.84FTIHH110 pKa = 6.35ILAAVAEE117 pKa = 4.41DD118 pKa = 3.18NRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84ISEE125 pKa = 4.23TTKK128 pKa = 10.84DD129 pKa = 3.37ALGAAKK135 pKa = 10.15RR136 pKa = 11.84RR137 pKa = 11.84GVILGKK143 pKa = 10.36NGTALSIANKK153 pKa = 9.73KK154 pKa = 9.32AAEE157 pKa = 4.42DD158 pKa = 3.85FAQQLSPTLKK168 pKa = 9.69RR169 pKa = 11.84LKK171 pKa = 9.34TRR173 pKa = 11.84GIISVRR179 pKa = 11.84AVCRR183 pKa = 11.84EE184 pKa = 3.84LNKK187 pKa = 10.44RR188 pKa = 11.84QIPTFRR194 pKa = 11.84GDD196 pKa = 4.0GRR198 pKa = 11.84WHH200 pKa = 6.95PSTVNVLMNRR210 pKa = 11.84LKK212 pKa = 9.68QTIISPP218 pKa = 4.03
Molecular weight: 24.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2137984 |
26 |
4355 |
365.1 |
40.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.56 ± 0.03 | 0.805 ± 0.01 |
5.059 ± 0.031 | 5.843 ± 0.033 |
5.002 ± 0.024 | 7.006 ± 0.038 |
1.861 ± 0.016 | 6.471 ± 0.024 |
6.428 ± 0.036 | 9.663 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.23 ± 0.015 | 5.238 ± 0.032 |
4.193 ± 0.019 | 4.034 ± 0.019 |
4.412 ± 0.025 | 6.147 ± 0.024 |
6.111 ± 0.043 | 6.634 ± 0.025 |
1.368 ± 0.014 | 3.934 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
