
Eubacterium siraeum CAG:80
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; environmental samples
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
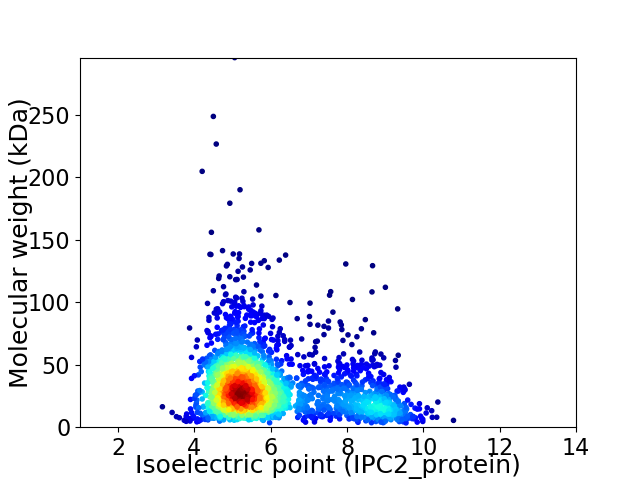
Virtual 2D-PAGE plot for 2308 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6R3J8|R6R3J8_9FIRM Type IV secretory pathway VirD4 components OS=Eubacterium siraeum CAG:80 OX=1263080 GN=BN788_00046 PE=3 SV=1
MM1 pKa = 7.95RR2 pKa = 11.84KK3 pKa = 7.73TSEE6 pKa = 4.18KK7 pKa = 10.18ILRR10 pKa = 11.84ALLAVSVLATAVLSSGCLDD29 pKa = 3.9DD30 pKa = 5.22EE31 pKa = 4.87EE32 pKa = 5.26EE33 pKa = 4.71ISDD36 pKa = 3.97STASDD41 pKa = 3.18NRR43 pKa = 11.84QTSDD47 pKa = 2.76GGNAEE52 pKa = 4.11SSEE55 pKa = 4.14GSDD58 pKa = 2.89NKK60 pKa = 10.74YY61 pKa = 10.64DD62 pKa = 3.69GSVTGSRR69 pKa = 11.84ASKK72 pKa = 9.15LTFSGSDD79 pKa = 3.62GISIARR85 pKa = 11.84KK86 pKa = 8.07QRR88 pKa = 11.84EE89 pKa = 4.09AEE91 pKa = 4.03KK92 pKa = 10.89PMGEE96 pKa = 5.21DD97 pKa = 3.04GTQTVFVYY105 pKa = 9.57MCGSDD110 pKa = 4.03LEE112 pKa = 4.68SEE114 pKa = 4.38NGLASGDD121 pKa = 3.83IEE123 pKa = 4.42EE124 pKa = 5.18MIAGSQSEE132 pKa = 4.17NVKK135 pKa = 10.24FVIQTGGAGAWADD148 pKa = 3.63TYY150 pKa = 11.3GISAEE155 pKa = 3.95KK156 pKa = 7.26TQRR159 pKa = 11.84YY160 pKa = 7.45VVTGGEE166 pKa = 3.47ISLIEE171 pKa = 4.2EE172 pKa = 4.3KK173 pKa = 10.85EE174 pKa = 4.23SVNMGKK180 pKa = 10.15EE181 pKa = 3.87DD182 pKa = 3.61VLVDD186 pKa = 3.75FLGWGIEE193 pKa = 4.02NYY195 pKa = 10.13AAAKK199 pKa = 8.98MGLIFWNHH207 pKa = 5.31GGGSISGVCFDD218 pKa = 5.1EE219 pKa = 5.85LNEE222 pKa = 4.12NDD224 pKa = 4.08SLSLEE229 pKa = 4.69EE230 pKa = 5.23IDD232 pKa = 3.98TALTSIYY239 pKa = 11.13DD240 pKa = 3.37KK241 pKa = 9.39MTDD244 pKa = 2.76KK245 pKa = 10.84FAFIGFDD252 pKa = 3.37ACLMATVEE260 pKa = 4.37TANMLVPHH268 pKa = 7.5ADD270 pKa = 3.15YY271 pKa = 10.39MFASEE276 pKa = 4.35EE277 pKa = 4.3TEE279 pKa = 4.27PGYY282 pKa = 11.15GWDD285 pKa = 3.57YY286 pKa = 11.37TEE288 pKa = 4.12IAGFMEE294 pKa = 4.95SNPTADD300 pKa = 3.26TAEE303 pKa = 4.28LGKK306 pKa = 8.6TVADD310 pKa = 3.96SFMASCEE317 pKa = 4.11AIGAGGEE324 pKa = 4.18ATLSITDD331 pKa = 3.96LSRR334 pKa = 11.84IDD336 pKa = 4.09EE337 pKa = 4.27LVKK340 pKa = 10.75AVNDD344 pKa = 3.69AAEE347 pKa = 4.13EE348 pKa = 4.11MNDD351 pKa = 3.3ISSDD355 pKa = 3.57PALLANAVRR364 pKa = 11.84SIYY367 pKa = 7.69TVRR370 pKa = 11.84AYY372 pKa = 10.58GSNNDD377 pKa = 3.54TEE379 pKa = 6.07GYY381 pKa = 9.47TNMVDD386 pKa = 3.79LGSMIAATVSGDD398 pKa = 3.34KK399 pKa = 10.49ADD401 pKa = 3.91KK402 pKa = 11.08VMSALEE408 pKa = 3.89KK409 pKa = 10.25AVVYY413 pKa = 10.48NIYY416 pKa = 11.01GEE418 pKa = 4.89DD419 pKa = 3.67KK420 pKa = 10.98DD421 pKa = 4.55GSTGLSTYY429 pKa = 10.71YY430 pKa = 10.09PFGVSGSAEE439 pKa = 3.76LATFGKK445 pKa = 10.43VCVSPYY451 pKa = 11.2YY452 pKa = 8.6MTFVDD457 pKa = 5.74RR458 pKa = 11.84IAYY461 pKa = 7.64GAQNGSTDD469 pKa = 3.46GYY471 pKa = 10.54SGEE474 pKa = 4.4KK475 pKa = 8.62TWLADD480 pKa = 3.55GAVYY484 pKa = 10.06WSDD487 pKa = 3.18GDD489 pKa = 3.85YY490 pKa = 11.37SDD492 pKa = 5.93YY493 pKa = 10.96EE494 pKa = 4.98SNWEE498 pKa = 4.07HH499 pKa = 6.54YY500 pKa = 10.33NNKK503 pKa = 7.51TDD505 pKa = 4.07NMGFCGDD512 pKa = 3.79KK513 pKa = 10.86SSVQIDD519 pKa = 3.68VGPVLDD525 pKa = 5.52DD526 pKa = 3.81EE527 pKa = 4.89GTFYY531 pKa = 11.33FNVTDD536 pKa = 3.65DD537 pKa = 4.51TINNLASVNCSVYY550 pKa = 10.49RR551 pKa = 11.84VDD553 pKa = 4.54EE554 pKa = 4.26EE555 pKa = 4.17TGEE558 pKa = 4.3LIEE561 pKa = 5.1FGVDD565 pKa = 2.93QSTKK569 pKa = 9.92VDD571 pKa = 4.53FSSGLVEE578 pKa = 6.25DD579 pKa = 4.38NFSGQWYY586 pKa = 8.82MIEE589 pKa = 4.21NNLIPMFIVEE599 pKa = 4.56KK600 pKa = 10.44NGNSSVYY607 pKa = 9.58TSPVKK612 pKa = 10.99LNGKK616 pKa = 6.55EE617 pKa = 4.1TNLRR621 pKa = 11.84IAMTQDD627 pKa = 2.43GNAYY631 pKa = 10.25DD632 pKa = 3.77ITALGTWDD640 pKa = 3.44GVNEE644 pKa = 4.16NGEE647 pKa = 4.41SARR650 pKa = 11.84SVVPLKK656 pKa = 10.61EE657 pKa = 3.74GDD659 pKa = 3.79VIVPIFNTYY668 pKa = 10.71DD669 pKa = 3.4SEE671 pKa = 5.09GNFAGKK677 pKa = 10.48AEE679 pKa = 4.27GDD681 pKa = 3.51EE682 pKa = 4.44CTYY685 pKa = 11.24SGDD688 pKa = 3.76NMIEE692 pKa = 4.17FVNLPAGDD700 pKa = 3.69YY701 pKa = 10.2RR702 pKa = 11.84YY703 pKa = 10.97SFVINDD709 pKa = 3.28IYY711 pKa = 11.6GNVCYY716 pKa = 10.03TGFTVFTTDD725 pKa = 3.59DD726 pKa = 3.97DD727 pKa = 5.22GNVFFTPEE735 pKa = 3.56EE736 pKa = 4.0
MM1 pKa = 7.95RR2 pKa = 11.84KK3 pKa = 7.73TSEE6 pKa = 4.18KK7 pKa = 10.18ILRR10 pKa = 11.84ALLAVSVLATAVLSSGCLDD29 pKa = 3.9DD30 pKa = 5.22EE31 pKa = 4.87EE32 pKa = 5.26EE33 pKa = 4.71ISDD36 pKa = 3.97STASDD41 pKa = 3.18NRR43 pKa = 11.84QTSDD47 pKa = 2.76GGNAEE52 pKa = 4.11SSEE55 pKa = 4.14GSDD58 pKa = 2.89NKK60 pKa = 10.74YY61 pKa = 10.64DD62 pKa = 3.69GSVTGSRR69 pKa = 11.84ASKK72 pKa = 9.15LTFSGSDD79 pKa = 3.62GISIARR85 pKa = 11.84KK86 pKa = 8.07QRR88 pKa = 11.84EE89 pKa = 4.09AEE91 pKa = 4.03KK92 pKa = 10.89PMGEE96 pKa = 5.21DD97 pKa = 3.04GTQTVFVYY105 pKa = 9.57MCGSDD110 pKa = 4.03LEE112 pKa = 4.68SEE114 pKa = 4.38NGLASGDD121 pKa = 3.83IEE123 pKa = 4.42EE124 pKa = 5.18MIAGSQSEE132 pKa = 4.17NVKK135 pKa = 10.24FVIQTGGAGAWADD148 pKa = 3.63TYY150 pKa = 11.3GISAEE155 pKa = 3.95KK156 pKa = 7.26TQRR159 pKa = 11.84YY160 pKa = 7.45VVTGGEE166 pKa = 3.47ISLIEE171 pKa = 4.2EE172 pKa = 4.3KK173 pKa = 10.85EE174 pKa = 4.23SVNMGKK180 pKa = 10.15EE181 pKa = 3.87DD182 pKa = 3.61VLVDD186 pKa = 3.75FLGWGIEE193 pKa = 4.02NYY195 pKa = 10.13AAAKK199 pKa = 8.98MGLIFWNHH207 pKa = 5.31GGGSISGVCFDD218 pKa = 5.1EE219 pKa = 5.85LNEE222 pKa = 4.12NDD224 pKa = 4.08SLSLEE229 pKa = 4.69EE230 pKa = 5.23IDD232 pKa = 3.98TALTSIYY239 pKa = 11.13DD240 pKa = 3.37KK241 pKa = 9.39MTDD244 pKa = 2.76KK245 pKa = 10.84FAFIGFDD252 pKa = 3.37ACLMATVEE260 pKa = 4.37TANMLVPHH268 pKa = 7.5ADD270 pKa = 3.15YY271 pKa = 10.39MFASEE276 pKa = 4.35EE277 pKa = 4.3TEE279 pKa = 4.27PGYY282 pKa = 11.15GWDD285 pKa = 3.57YY286 pKa = 11.37TEE288 pKa = 4.12IAGFMEE294 pKa = 4.95SNPTADD300 pKa = 3.26TAEE303 pKa = 4.28LGKK306 pKa = 8.6TVADD310 pKa = 3.96SFMASCEE317 pKa = 4.11AIGAGGEE324 pKa = 4.18ATLSITDD331 pKa = 3.96LSRR334 pKa = 11.84IDD336 pKa = 4.09EE337 pKa = 4.27LVKK340 pKa = 10.75AVNDD344 pKa = 3.69AAEE347 pKa = 4.13EE348 pKa = 4.11MNDD351 pKa = 3.3ISSDD355 pKa = 3.57PALLANAVRR364 pKa = 11.84SIYY367 pKa = 7.69TVRR370 pKa = 11.84AYY372 pKa = 10.58GSNNDD377 pKa = 3.54TEE379 pKa = 6.07GYY381 pKa = 9.47TNMVDD386 pKa = 3.79LGSMIAATVSGDD398 pKa = 3.34KK399 pKa = 10.49ADD401 pKa = 3.91KK402 pKa = 11.08VMSALEE408 pKa = 3.89KK409 pKa = 10.25AVVYY413 pKa = 10.48NIYY416 pKa = 11.01GEE418 pKa = 4.89DD419 pKa = 3.67KK420 pKa = 10.98DD421 pKa = 4.55GSTGLSTYY429 pKa = 10.71YY430 pKa = 10.09PFGVSGSAEE439 pKa = 3.76LATFGKK445 pKa = 10.43VCVSPYY451 pKa = 11.2YY452 pKa = 8.6MTFVDD457 pKa = 5.74RR458 pKa = 11.84IAYY461 pKa = 7.64GAQNGSTDD469 pKa = 3.46GYY471 pKa = 10.54SGEE474 pKa = 4.4KK475 pKa = 8.62TWLADD480 pKa = 3.55GAVYY484 pKa = 10.06WSDD487 pKa = 3.18GDD489 pKa = 3.85YY490 pKa = 11.37SDD492 pKa = 5.93YY493 pKa = 10.96EE494 pKa = 4.98SNWEE498 pKa = 4.07HH499 pKa = 6.54YY500 pKa = 10.33NNKK503 pKa = 7.51TDD505 pKa = 4.07NMGFCGDD512 pKa = 3.79KK513 pKa = 10.86SSVQIDD519 pKa = 3.68VGPVLDD525 pKa = 5.52DD526 pKa = 3.81EE527 pKa = 4.89GTFYY531 pKa = 11.33FNVTDD536 pKa = 3.65DD537 pKa = 4.51TINNLASVNCSVYY550 pKa = 10.49RR551 pKa = 11.84VDD553 pKa = 4.54EE554 pKa = 4.26EE555 pKa = 4.17TGEE558 pKa = 4.3LIEE561 pKa = 5.1FGVDD565 pKa = 2.93QSTKK569 pKa = 9.92VDD571 pKa = 4.53FSSGLVEE578 pKa = 6.25DD579 pKa = 4.38NFSGQWYY586 pKa = 8.82MIEE589 pKa = 4.21NNLIPMFIVEE599 pKa = 4.56KK600 pKa = 10.44NGNSSVYY607 pKa = 9.58TSPVKK612 pKa = 10.99LNGKK616 pKa = 6.55EE617 pKa = 4.1TNLRR621 pKa = 11.84IAMTQDD627 pKa = 2.43GNAYY631 pKa = 10.25DD632 pKa = 3.77ITALGTWDD640 pKa = 3.44GVNEE644 pKa = 4.16NGEE647 pKa = 4.41SARR650 pKa = 11.84SVVPLKK656 pKa = 10.61EE657 pKa = 3.74GDD659 pKa = 3.79VIVPIFNTYY668 pKa = 10.71DD669 pKa = 3.4SEE671 pKa = 5.09GNFAGKK677 pKa = 10.48AEE679 pKa = 4.27GDD681 pKa = 3.51EE682 pKa = 4.44CTYY685 pKa = 11.24SGDD688 pKa = 3.76NMIEE692 pKa = 4.17FVNLPAGDD700 pKa = 3.69YY701 pKa = 10.2RR702 pKa = 11.84YY703 pKa = 10.97SFVINDD709 pKa = 3.28IYY711 pKa = 11.6GNVCYY716 pKa = 10.03TGFTVFTTDD725 pKa = 3.59DD726 pKa = 3.97DD727 pKa = 5.22GNVFFTPEE735 pKa = 3.56EE736 pKa = 4.0
Molecular weight: 79.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6RFY1|R6RFY1_9FIRM DUF4430 domain-containing protein OS=Eubacterium siraeum CAG:80 OX=1263080 GN=BN788_01428 PE=4 SV=1
MM1 pKa = 6.78NRR3 pKa = 11.84KK4 pKa = 7.4MKK6 pKa = 9.94KK7 pKa = 8.41YY8 pKa = 10.27RR9 pKa = 11.84YY10 pKa = 9.37IISKK14 pKa = 9.28NKK16 pKa = 9.58KK17 pKa = 7.13VQLYY21 pKa = 8.29HH22 pKa = 6.47ICGCGGCRR30 pKa = 11.84CIRR33 pKa = 11.84NFIRR37 pKa = 11.84LHH39 pKa = 5.17IRR41 pKa = 11.84QGEE44 pKa = 4.09QCVICRR50 pKa = 11.84TGAFDD55 pKa = 5.0FGSGKK60 pKa = 9.6CRR62 pKa = 11.84KK63 pKa = 9.76LGGRR67 pKa = 11.84YY68 pKa = 8.94KK69 pKa = 10.82RR70 pKa = 11.84LGKK73 pKa = 10.31
MM1 pKa = 6.78NRR3 pKa = 11.84KK4 pKa = 7.4MKK6 pKa = 9.94KK7 pKa = 8.41YY8 pKa = 10.27RR9 pKa = 11.84YY10 pKa = 9.37IISKK14 pKa = 9.28NKK16 pKa = 9.58KK17 pKa = 7.13VQLYY21 pKa = 8.29HH22 pKa = 6.47ICGCGGCRR30 pKa = 11.84CIRR33 pKa = 11.84NFIRR37 pKa = 11.84LHH39 pKa = 5.17IRR41 pKa = 11.84QGEE44 pKa = 4.09QCVICRR50 pKa = 11.84TGAFDD55 pKa = 5.0FGSGKK60 pKa = 9.6CRR62 pKa = 11.84KK63 pKa = 9.76LGGRR67 pKa = 11.84YY68 pKa = 8.94KK69 pKa = 10.82RR70 pKa = 11.84LGKK73 pKa = 10.31
Molecular weight: 8.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
734533 |
29 |
2601 |
318.3 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.81 ± 0.046 | 1.747 ± 0.024 |
6.317 ± 0.041 | 6.694 ± 0.046 |
4.123 ± 0.037 | 6.961 ± 0.05 |
1.475 ± 0.018 | 7.595 ± 0.047 |
7.18 ± 0.048 | 8.211 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.815 ± 0.029 | 4.844 ± 0.042 |
3.207 ± 0.03 | 2.571 ± 0.027 |
4.029 ± 0.044 | 6.573 ± 0.052 |
6.044 ± 0.052 | 6.841 ± 0.041 |
0.755 ± 0.016 | 4.204 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
