
Cellulomonas sp. WB94
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; unclassified Cellulomonas
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
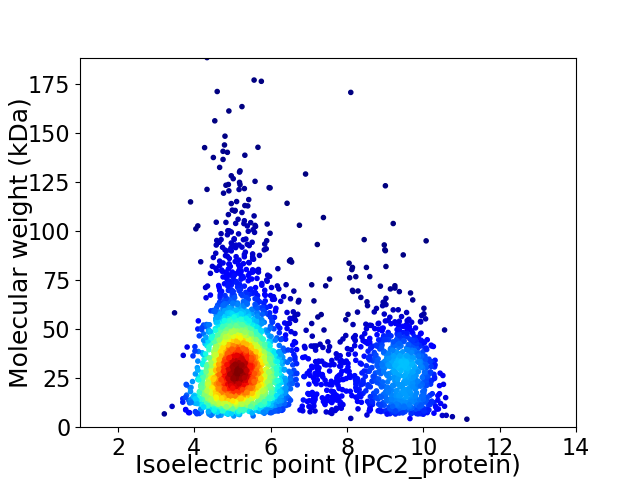
Virtual 2D-PAGE plot for 3376 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
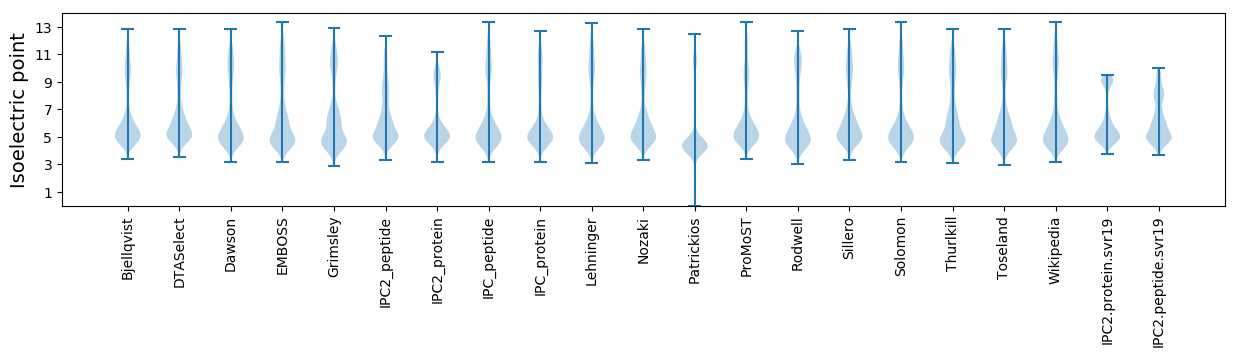
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T9XMW1|A0A2T9XMW1_9CELL Redoxin OS=Cellulomonas sp. WB94 OX=2173174 GN=DDP54_16035 PE=4 SV=1
MM1 pKa = 8.02GDD3 pKa = 3.51PSLPGPMRR11 pKa = 11.84RR12 pKa = 11.84IPVKK16 pKa = 10.49LSWRR20 pKa = 11.84VGATMLAVAAVVAVPVPALADD41 pKa = 3.64TVAVTNAVVAGDD53 pKa = 3.7GSKK56 pKa = 10.33SVRR59 pKa = 11.84TDD61 pKa = 2.83TGVGVTVSYY70 pKa = 10.04TLTAEE75 pKa = 4.06NNPVGDD81 pKa = 3.97LQGCNAQTKK90 pKa = 8.71PGGGVTVTILPAANVTASPSALIFTDD116 pKa = 4.08CSTAQSVTFSASSPGDD132 pKa = 3.5YY133 pKa = 10.62SVTSSATGGKK143 pKa = 7.88TGSLFTTTASNFTLHH158 pKa = 6.16VVSPNQPPTVTVSGVLQGASYY179 pKa = 10.76EE180 pKa = 4.36HH181 pKa = 6.84GSVPAAMCNVTDD193 pKa = 3.8DD194 pKa = 4.22HH195 pKa = 7.4DD196 pKa = 3.97VARR199 pKa = 11.84TFAATLSAITGTLTSYY215 pKa = 11.61GLGGRR220 pKa = 11.84TATCAATDD228 pKa = 3.69TGGLTSTTAASYY240 pKa = 9.45TIVDD244 pKa = 3.7TTRR247 pKa = 11.84PILTVPGDD255 pKa = 3.86LTAEE259 pKa = 4.24ATSSSGAVVTWSASATDD276 pKa = 4.21AVDD279 pKa = 4.0GSPAVACSGAGGLTSGATFPLGATLVGCTATDD311 pKa = 3.46VAGNQATQQFQVTVQDD327 pKa = 3.64TTPPLLTLPASSTTEE342 pKa = 3.56ATGPSGASVTYY353 pKa = 10.14SASATDD359 pKa = 4.4SVDD362 pKa = 3.48GDD364 pKa = 3.98LTPTCTPASATTFGITTTTVDD385 pKa = 3.49CTATDD390 pKa = 2.95AHH392 pKa = 6.77ANHH395 pKa = 6.51AQGLFSVTVQDD406 pKa = 3.69TTPPTLTLPTVAPRR420 pKa = 11.84EE421 pKa = 4.22ATGPSGATVTYY432 pKa = 10.19AATASDD438 pKa = 4.32VVDD441 pKa = 3.38LTVPVTCDD449 pKa = 3.26YY450 pKa = 7.89PTGSVFQVGDD460 pKa = 3.65TTVSCTATDD469 pKa = 3.23SHH471 pKa = 7.08GNTTPGHH478 pKa = 5.06FTVTVTDD485 pKa = 3.36TTGPVLTLPGNISEE499 pKa = 4.78EE500 pKa = 4.34ATKK503 pKa = 9.85STGSDD508 pKa = 2.4IAYY511 pKa = 10.29SVLASDD517 pKa = 5.29LVDD520 pKa = 3.39GTVATSCDD528 pKa = 3.46PANGTFAIGTTTVTCTATDD547 pKa = 3.05AHH549 pKa = 6.92ANTTTGSFTVTVVDD563 pKa = 3.72TTAPVFSAGPGDD575 pKa = 3.84QTLEE579 pKa = 3.8AAGADD584 pKa = 3.55GTVATFASPTAVDD597 pKa = 4.71LVDD600 pKa = 3.56GDD602 pKa = 4.1RR603 pKa = 11.84AVLCVSSSGLGSGSTFPLGTTTVTCVASDD632 pKa = 3.36TRR634 pKa = 11.84GNVSDD639 pKa = 3.3QTFTVKK645 pKa = 10.39VSDD648 pKa = 4.4TIAPLVGVLEE658 pKa = 4.22NLTPEE663 pKa = 3.85ATGPDD668 pKa = 3.62GAVVSFTAPTATDD681 pKa = 3.46AVSGSPAVTCDD692 pKa = 3.41PASGSTFAIDD702 pKa = 3.32TVATITCSATDD713 pKa = 3.25SSRR716 pKa = 11.84NTGSSTFTVVVHH728 pKa = 7.21DD729 pKa = 3.89STGPALAVPANITEE743 pKa = 4.45EE744 pKa = 4.13ATSPDD749 pKa = 3.14GAAASWTDD757 pKa = 3.31ASATDD762 pKa = 4.62LVDD765 pKa = 4.25GPLPATCDD773 pKa = 3.49PASGSVFPLGNATAVTCTAYY793 pKa = 10.25DD794 pKa = 3.46VRR796 pKa = 11.84HH797 pKa = 5.73NKK799 pKa = 9.52GVGGFTVTVLDD810 pKa = 3.84RR811 pKa = 11.84TAPTLAVSGDD821 pKa = 3.65SALEE825 pKa = 3.73ATAPSGAPATFTATATDD842 pKa = 3.84LVDD845 pKa = 4.32GVVTPTCTAGGDD857 pKa = 4.0PVASGATFPLGTTTVTCSATDD878 pKa = 3.18SHH880 pKa = 6.89GNNAPSQSFTITVQDD895 pKa = 3.41TTRR898 pKa = 11.84PSVTVPATMTLEE910 pKa = 4.08ATSANGAPGTFASSATDD927 pKa = 3.63LVDD930 pKa = 3.51GTLTPTCTPASGATFPLGATTVTCSATDD958 pKa = 3.13AHH960 pKa = 7.05ANTATGSFAVTVVDD974 pKa = 3.74TTAPVVSVPANVATFATSAAGAVVTFAAPTASDD1007 pKa = 3.44AVGGNSLAVCTPPSGSLFKK1026 pKa = 10.96PGDD1029 pKa = 3.82TVVSCTATDD1038 pKa = 3.41AAGNSASRR1046 pKa = 11.84TFKK1049 pKa = 10.94VIVRR1053 pKa = 11.84FDD1055 pKa = 3.27WTGFFAPVDD1064 pKa = 3.72NAILNGMKK1072 pKa = 10.2AGSTAPIKK1080 pKa = 9.91WQVSNQGGGYY1090 pKa = 10.05VPDD1093 pKa = 3.56LTIVSKK1099 pKa = 7.34TTSGIAICATGALVDD1114 pKa = 4.32DD1115 pKa = 4.97LEE1117 pKa = 5.66AYY1119 pKa = 8.06STGGTQLRR1127 pKa = 11.84YY1128 pKa = 10.49DD1129 pKa = 3.75STAHH1133 pKa = 4.81QFIYY1137 pKa = 10.34NWQSPKK1143 pKa = 10.68KK1144 pKa = 9.87PGTCYY1149 pKa = 10.36TIIIGLTDD1157 pKa = 3.8GTSHH1161 pKa = 7.22SATFQLKK1168 pKa = 9.86
MM1 pKa = 8.02GDD3 pKa = 3.51PSLPGPMRR11 pKa = 11.84RR12 pKa = 11.84IPVKK16 pKa = 10.49LSWRR20 pKa = 11.84VGATMLAVAAVVAVPVPALADD41 pKa = 3.64TVAVTNAVVAGDD53 pKa = 3.7GSKK56 pKa = 10.33SVRR59 pKa = 11.84TDD61 pKa = 2.83TGVGVTVSYY70 pKa = 10.04TLTAEE75 pKa = 4.06NNPVGDD81 pKa = 3.97LQGCNAQTKK90 pKa = 8.71PGGGVTVTILPAANVTASPSALIFTDD116 pKa = 4.08CSTAQSVTFSASSPGDD132 pKa = 3.5YY133 pKa = 10.62SVTSSATGGKK143 pKa = 7.88TGSLFTTTASNFTLHH158 pKa = 6.16VVSPNQPPTVTVSGVLQGASYY179 pKa = 10.76EE180 pKa = 4.36HH181 pKa = 6.84GSVPAAMCNVTDD193 pKa = 3.8DD194 pKa = 4.22HH195 pKa = 7.4DD196 pKa = 3.97VARR199 pKa = 11.84TFAATLSAITGTLTSYY215 pKa = 11.61GLGGRR220 pKa = 11.84TATCAATDD228 pKa = 3.69TGGLTSTTAASYY240 pKa = 9.45TIVDD244 pKa = 3.7TTRR247 pKa = 11.84PILTVPGDD255 pKa = 3.86LTAEE259 pKa = 4.24ATSSSGAVVTWSASATDD276 pKa = 4.21AVDD279 pKa = 4.0GSPAVACSGAGGLTSGATFPLGATLVGCTATDD311 pKa = 3.46VAGNQATQQFQVTVQDD327 pKa = 3.64TTPPLLTLPASSTTEE342 pKa = 3.56ATGPSGASVTYY353 pKa = 10.14SASATDD359 pKa = 4.4SVDD362 pKa = 3.48GDD364 pKa = 3.98LTPTCTPASATTFGITTTTVDD385 pKa = 3.49CTATDD390 pKa = 2.95AHH392 pKa = 6.77ANHH395 pKa = 6.51AQGLFSVTVQDD406 pKa = 3.69TTPPTLTLPTVAPRR420 pKa = 11.84EE421 pKa = 4.22ATGPSGATVTYY432 pKa = 10.19AATASDD438 pKa = 4.32VVDD441 pKa = 3.38LTVPVTCDD449 pKa = 3.26YY450 pKa = 7.89PTGSVFQVGDD460 pKa = 3.65TTVSCTATDD469 pKa = 3.23SHH471 pKa = 7.08GNTTPGHH478 pKa = 5.06FTVTVTDD485 pKa = 3.36TTGPVLTLPGNISEE499 pKa = 4.78EE500 pKa = 4.34ATKK503 pKa = 9.85STGSDD508 pKa = 2.4IAYY511 pKa = 10.29SVLASDD517 pKa = 5.29LVDD520 pKa = 3.39GTVATSCDD528 pKa = 3.46PANGTFAIGTTTVTCTATDD547 pKa = 3.05AHH549 pKa = 6.92ANTTTGSFTVTVVDD563 pKa = 3.72TTAPVFSAGPGDD575 pKa = 3.84QTLEE579 pKa = 3.8AAGADD584 pKa = 3.55GTVATFASPTAVDD597 pKa = 4.71LVDD600 pKa = 3.56GDD602 pKa = 4.1RR603 pKa = 11.84AVLCVSSSGLGSGSTFPLGTTTVTCVASDD632 pKa = 3.36TRR634 pKa = 11.84GNVSDD639 pKa = 3.3QTFTVKK645 pKa = 10.39VSDD648 pKa = 4.4TIAPLVGVLEE658 pKa = 4.22NLTPEE663 pKa = 3.85ATGPDD668 pKa = 3.62GAVVSFTAPTATDD681 pKa = 3.46AVSGSPAVTCDD692 pKa = 3.41PASGSTFAIDD702 pKa = 3.32TVATITCSATDD713 pKa = 3.25SSRR716 pKa = 11.84NTGSSTFTVVVHH728 pKa = 7.21DD729 pKa = 3.89STGPALAVPANITEE743 pKa = 4.45EE744 pKa = 4.13ATSPDD749 pKa = 3.14GAAASWTDD757 pKa = 3.31ASATDD762 pKa = 4.62LVDD765 pKa = 4.25GPLPATCDD773 pKa = 3.49PASGSVFPLGNATAVTCTAYY793 pKa = 10.25DD794 pKa = 3.46VRR796 pKa = 11.84HH797 pKa = 5.73NKK799 pKa = 9.52GVGGFTVTVLDD810 pKa = 3.84RR811 pKa = 11.84TAPTLAVSGDD821 pKa = 3.65SALEE825 pKa = 3.73ATAPSGAPATFTATATDD842 pKa = 3.84LVDD845 pKa = 4.32GVVTPTCTAGGDD857 pKa = 4.0PVASGATFPLGTTTVTCSATDD878 pKa = 3.18SHH880 pKa = 6.89GNNAPSQSFTITVQDD895 pKa = 3.41TTRR898 pKa = 11.84PSVTVPATMTLEE910 pKa = 4.08ATSANGAPGTFASSATDD927 pKa = 3.63LVDD930 pKa = 3.51GTLTPTCTPASGATFPLGATTVTCSATDD958 pKa = 3.13AHH960 pKa = 7.05ANTATGSFAVTVVDD974 pKa = 3.74TTAPVVSVPANVATFATSAAGAVVTFAAPTASDD1007 pKa = 3.44AVGGNSLAVCTPPSGSLFKK1026 pKa = 10.96PGDD1029 pKa = 3.82TVVSCTATDD1038 pKa = 3.41AAGNSASRR1046 pKa = 11.84TFKK1049 pKa = 10.94VIVRR1053 pKa = 11.84FDD1055 pKa = 3.27WTGFFAPVDD1064 pKa = 3.72NAILNGMKK1072 pKa = 10.2AGSTAPIKK1080 pKa = 9.91WQVSNQGGGYY1090 pKa = 10.05VPDD1093 pKa = 3.56LTIVSKK1099 pKa = 7.34TTSGIAICATGALVDD1114 pKa = 4.32DD1115 pKa = 4.97LEE1117 pKa = 5.66AYY1119 pKa = 8.06STGGTQLRR1127 pKa = 11.84YY1128 pKa = 10.49DD1129 pKa = 3.75STAHH1133 pKa = 4.81QFIYY1137 pKa = 10.34NWQSPKK1143 pKa = 10.68KK1144 pKa = 9.87PGTCYY1149 pKa = 10.36TIIIGLTDD1157 pKa = 3.8GTSHH1161 pKa = 7.22SATFQLKK1168 pKa = 9.86
Molecular weight: 115.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T9XQX5|A0A2T9XQX5_9CELL DUF5666 domain-containing protein OS=Cellulomonas sp. WB94 OX=2173174 GN=DDP54_05220 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1112575 |
32 |
1819 |
329.6 |
34.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.233 ± 0.067 | 0.599 ± 0.011 |
6.209 ± 0.033 | 4.908 ± 0.032 |
2.636 ± 0.025 | 9.245 ± 0.039 |
2.124 ± 0.021 | 3.541 ± 0.026 |
1.606 ± 0.027 | 10.376 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.018 | 1.642 ± 0.022 |
5.707 ± 0.036 | 2.692 ± 0.021 |
7.604 ± 0.05 | 5.416 ± 0.03 |
6.509 ± 0.038 | 9.937 ± 0.04 |
1.488 ± 0.018 | 1.844 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
