
Xenopus tropicalis (Western clawed frog) (Silurana tropicalis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amphibia; Batrachia; Anura; Pipoidea
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
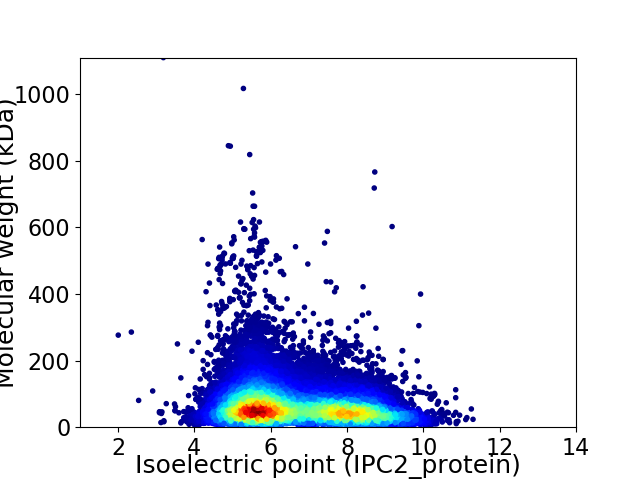
Virtual 2D-PAGE plot for 46313 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A803K5Q7|A0A803K5Q7_XENTR F-box/WD repeat-containing protein 7-like OS=Xenopus tropicalis OX=8364 GN=LOC116410231 PE=4 SV=1
KKK2 pKa = 10.14DD3 pKa = 3.63PTPRR7 pKa = 11.84CLCNQSIGWCRR18 pKa = 11.84LYYY21 pKa = 10.97VNPCCEEE28 pKa = 4.03ALSLVVCSYYY38 pKa = 10.23GQKKK42 pKa = 7.44TQIYYY47 pKa = 8.51GQKKK51 pKa = 9.05PQIYYY56 pKa = 8.75GQKKK60 pKa = 8.98PQIYYY65 pKa = 8.75GQKKK69 pKa = 6.43TQIYYY74 pKa = 8.47GQKKK78 pKa = 6.45TQIYYY83 pKa = 8.47GQKKK87 pKa = 6.45TQIYYY92 pKa = 8.47GQKKK96 pKa = 6.45TQIYYY101 pKa = 8.47GQKKK105 pKa = 6.45TQIYYY110 pKa = 8.47GQKKK114 pKa = 6.45TQIYYY119 pKa = 8.47GQKKK123 pKa = 5.86TQIFTGQKKK132 pKa = 10.03EEE134 pKa = 4.1DDD136 pKa = 4.3DDD138 pKa = 3.63HHH140 pKa = 6.71PEEE143 pKa = 4.2DDD145 pKa = 4.92DDD147 pKa = 3.77HHH149 pKa = 6.71PEEE152 pKa = 4.2DDD154 pKa = 4.92DDD156 pKa = 3.77HHH158 pKa = 6.71PEEE161 pKa = 4.2DDD163 pKa = 4.92DDD165 pKa = 3.77HHH167 pKa = 6.71PEEE170 pKa = 4.2DDD172 pKa = 4.92DDD174 pKa = 3.77HHH176 pKa = 6.71PEEE179 pKa = 4.2DDD181 pKa = 4.92DDD183 pKa = 3.77HHH185 pKa = 6.71PEEE188 pKa = 4.2DDD190 pKa = 4.92DDD192 pKa = 3.77HHH194 pKa = 6.71PEEE197 pKa = 4.2DDD199 pKa = 4.92DDD201 pKa = 3.77HHH203 pKa = 6.71PEEE206 pKa = 4.2DDD208 pKa = 4.92DDD210 pKa = 3.77HHH212 pKa = 6.71PEEE215 pKa = 4.2DDD217 pKa = 4.92DDD219 pKa = 3.77HHH221 pKa = 6.71PEEE224 pKa = 4.2DDD226 pKa = 4.92DDD228 pKa = 3.77HHH230 pKa = 6.71PEEE233 pKa = 4.2DDD235 pKa = 4.92DDD237 pKa = 3.77HHH239 pKa = 6.71PEEE242 pKa = 4.2DDD244 pKa = 4.92DDD246 pKa = 3.77HHH248 pKa = 6.71PEEE251 pKa = 4.2DDD253 pKa = 4.92DDD255 pKa = 3.77HHH257 pKa = 6.71PEEE260 pKa = 4.2DDD262 pKa = 4.92DDD264 pKa = 3.77HHH266 pKa = 6.71PEEE269 pKa = 4.2DDD271 pKa = 4.92DDD273 pKa = 3.77HHH275 pKa = 6.71PEEE278 pKa = 4.2DDD280 pKa = 4.92DDD282 pKa = 3.77HHH284 pKa = 6.71PEEE287 pKa = 4.2DDD289 pKa = 4.92DDD291 pKa = 3.77HHH293 pKa = 6.71PEEE296 pKa = 4.2DDD298 pKa = 4.92DDD300 pKa = 3.77HHH302 pKa = 6.71PEEE305 pKa = 4.2DDD307 pKa = 4.92DDD309 pKa = 3.77HHH311 pKa = 6.71PEEE314 pKa = 4.2DDD316 pKa = 4.92DDD318 pKa = 3.77HHH320 pKa = 6.71PEEE323 pKa = 4.2DDD325 pKa = 4.92DDD327 pKa = 3.77HHH329 pKa = 6.71PEEE332 pKa = 4.2DDD334 pKa = 4.92DDD336 pKa = 3.77HHH338 pKa = 6.71PEEE341 pKa = 4.2DDD343 pKa = 4.92DDD345 pKa = 3.77HHH347 pKa = 6.71PEEE350 pKa = 4.2DDD352 pKa = 4.92DDD354 pKa = 3.77HHH356 pKa = 6.71PEEE359 pKa = 4.2DDD361 pKa = 4.92DDD363 pKa = 3.77HHH365 pKa = 6.71PEEE368 pKa = 4.2DDD370 pKa = 4.92DDD372 pKa = 3.77HHH374 pKa = 6.71PEEE377 pKa = 4.2DDD379 pKa = 5.3DDD381 pKa = 3.72HHH383 pKa = 6.43P
KKK2 pKa = 10.14DD3 pKa = 3.63PTPRR7 pKa = 11.84CLCNQSIGWCRR18 pKa = 11.84LYYY21 pKa = 10.97VNPCCEEE28 pKa = 4.03ALSLVVCSYYY38 pKa = 10.23GQKKK42 pKa = 7.44TQIYYY47 pKa = 8.51GQKKK51 pKa = 9.05PQIYYY56 pKa = 8.75GQKKK60 pKa = 8.98PQIYYY65 pKa = 8.75GQKKK69 pKa = 6.43TQIYYY74 pKa = 8.47GQKKK78 pKa = 6.45TQIYYY83 pKa = 8.47GQKKK87 pKa = 6.45TQIYYY92 pKa = 8.47GQKKK96 pKa = 6.45TQIYYY101 pKa = 8.47GQKKK105 pKa = 6.45TQIYYY110 pKa = 8.47GQKKK114 pKa = 6.45TQIYYY119 pKa = 8.47GQKKK123 pKa = 5.86TQIFTGQKKK132 pKa = 10.03EEE134 pKa = 4.1DDD136 pKa = 4.3DDD138 pKa = 3.63HHH140 pKa = 6.71PEEE143 pKa = 4.2DDD145 pKa = 4.92DDD147 pKa = 3.77HHH149 pKa = 6.71PEEE152 pKa = 4.2DDD154 pKa = 4.92DDD156 pKa = 3.77HHH158 pKa = 6.71PEEE161 pKa = 4.2DDD163 pKa = 4.92DDD165 pKa = 3.77HHH167 pKa = 6.71PEEE170 pKa = 4.2DDD172 pKa = 4.92DDD174 pKa = 3.77HHH176 pKa = 6.71PEEE179 pKa = 4.2DDD181 pKa = 4.92DDD183 pKa = 3.77HHH185 pKa = 6.71PEEE188 pKa = 4.2DDD190 pKa = 4.92DDD192 pKa = 3.77HHH194 pKa = 6.71PEEE197 pKa = 4.2DDD199 pKa = 4.92DDD201 pKa = 3.77HHH203 pKa = 6.71PEEE206 pKa = 4.2DDD208 pKa = 4.92DDD210 pKa = 3.77HHH212 pKa = 6.71PEEE215 pKa = 4.2DDD217 pKa = 4.92DDD219 pKa = 3.77HHH221 pKa = 6.71PEEE224 pKa = 4.2DDD226 pKa = 4.92DDD228 pKa = 3.77HHH230 pKa = 6.71PEEE233 pKa = 4.2DDD235 pKa = 4.92DDD237 pKa = 3.77HHH239 pKa = 6.71PEEE242 pKa = 4.2DDD244 pKa = 4.92DDD246 pKa = 3.77HHH248 pKa = 6.71PEEE251 pKa = 4.2DDD253 pKa = 4.92DDD255 pKa = 3.77HHH257 pKa = 6.71PEEE260 pKa = 4.2DDD262 pKa = 4.92DDD264 pKa = 3.77HHH266 pKa = 6.71PEEE269 pKa = 4.2DDD271 pKa = 4.92DDD273 pKa = 3.77HHH275 pKa = 6.71PEEE278 pKa = 4.2DDD280 pKa = 4.92DDD282 pKa = 3.77HHH284 pKa = 6.71PEEE287 pKa = 4.2DDD289 pKa = 4.92DDD291 pKa = 3.77HHH293 pKa = 6.71PEEE296 pKa = 4.2DDD298 pKa = 4.92DDD300 pKa = 3.77HHH302 pKa = 6.71PEEE305 pKa = 4.2DDD307 pKa = 4.92DDD309 pKa = 3.77HHH311 pKa = 6.71PEEE314 pKa = 4.2DDD316 pKa = 4.92DDD318 pKa = 3.77HHH320 pKa = 6.71PEEE323 pKa = 4.2DDD325 pKa = 4.92DDD327 pKa = 3.77HHH329 pKa = 6.71PEEE332 pKa = 4.2DDD334 pKa = 4.92DDD336 pKa = 3.77HHH338 pKa = 6.71PEEE341 pKa = 4.2DDD343 pKa = 4.92DDD345 pKa = 3.77HHH347 pKa = 6.71PEEE350 pKa = 4.2DDD352 pKa = 4.92DDD354 pKa = 3.77HHH356 pKa = 6.71PEEE359 pKa = 4.2DDD361 pKa = 4.92DDD363 pKa = 3.77HHH365 pKa = 6.71PEEE368 pKa = 4.2DDD370 pKa = 4.92DDD372 pKa = 3.77HHH374 pKa = 6.71PEEE377 pKa = 4.2DDD379 pKa = 5.3DDD381 pKa = 3.72HHH383 pKa = 6.43P
Molecular weight: 45.61 kDa
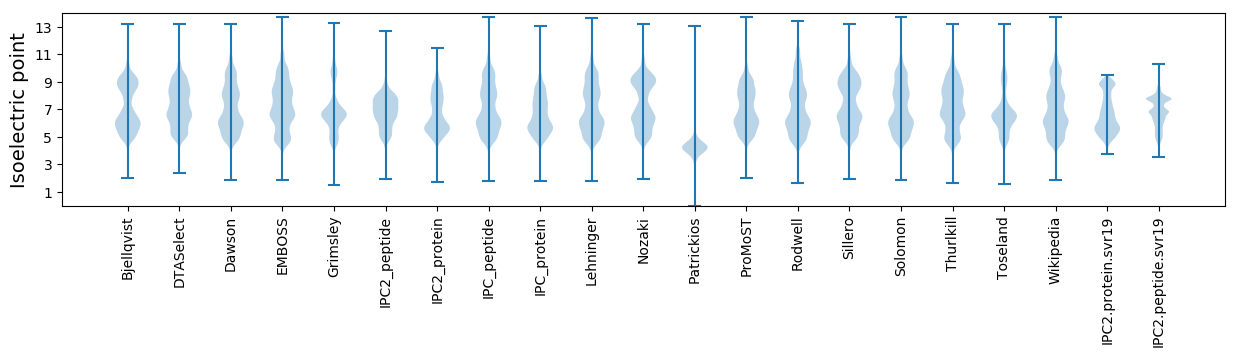
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A803JNG1|A0A803JNG1_XENTR Osteoclast stimulating factor 1 OS=Xenopus tropicalis OX=8364 GN=ostf1 PE=4 SV=1
MM1 pKa = 7.31TGYY4 pKa = 7.86TAAAVSVTPGKK15 pKa = 10.07CCRR18 pKa = 11.84GVHH21 pKa = 5.99KK22 pKa = 10.61LQARR26 pKa = 11.84GMHH29 pKa = 5.72KK30 pKa = 9.25QQARR34 pKa = 11.84GMHH37 pKa = 6.1KK38 pKa = 8.58PQARR42 pKa = 11.84GMHH45 pKa = 6.11KK46 pKa = 8.84PQARR50 pKa = 11.84GMHH53 pKa = 5.6KK54 pKa = 8.96QQARR58 pKa = 11.84GMHH61 pKa = 5.59KK62 pKa = 8.75QQARR66 pKa = 11.84GMHH69 pKa = 6.1KK70 pKa = 8.58PQARR74 pKa = 11.84GMHH77 pKa = 5.6KK78 pKa = 8.96QQARR82 pKa = 11.84GMHH85 pKa = 5.59KK86 pKa = 8.75QQARR90 pKa = 11.84GMHH93 pKa = 5.59KK94 pKa = 8.75QQARR98 pKa = 11.84GMHH101 pKa = 5.59KK102 pKa = 8.75QQARR106 pKa = 11.84GMHH109 pKa = 5.59KK110 pKa = 8.75QQARR114 pKa = 11.84GMHH117 pKa = 6.1KK118 pKa = 8.58PQARR122 pKa = 11.84GMHH125 pKa = 5.6KK126 pKa = 8.96QQARR130 pKa = 11.84GMHH133 pKa = 6.1KK134 pKa = 8.58PQARR138 pKa = 11.84GMHH141 pKa = 5.6KK142 pKa = 8.96QQARR146 pKa = 11.84GMHH149 pKa = 5.59KK150 pKa = 8.75QQARR154 pKa = 11.84GMHH157 pKa = 5.59KK158 pKa = 8.75QQARR162 pKa = 11.84GMHH165 pKa = 5.64KK166 pKa = 8.68QQARR170 pKa = 11.84GTHH173 pKa = 5.09KK174 pKa = 9.34QQARR178 pKa = 11.84GTHH181 pKa = 5.09KK182 pKa = 9.34QQARR186 pKa = 11.84GTHH189 pKa = 5.09KK190 pKa = 9.34QQARR194 pKa = 11.84GTHH197 pKa = 5.09KK198 pKa = 9.34QQARR202 pKa = 11.84GTHH205 pKa = 5.09KK206 pKa = 9.34QQARR210 pKa = 11.84GTHH213 pKa = 5.09KK214 pKa = 9.34QQARR218 pKa = 11.84GTHH221 pKa = 5.09KK222 pKa = 9.34QQARR226 pKa = 11.84GTHH229 pKa = 5.09KK230 pKa = 9.34QQARR234 pKa = 11.84GTHH237 pKa = 6.14KK238 pKa = 10.08LQARR242 pKa = 11.84GTHH245 pKa = 6.34KK246 pKa = 10.43LQARR250 pKa = 11.84GTHH253 pKa = 5.3KK254 pKa = 9.95QQARR258 pKa = 11.84GTHH261 pKa = 6.14KK262 pKa = 10.08LQARR266 pKa = 11.84GTHH269 pKa = 5.3KK270 pKa = 9.95QQARR274 pKa = 11.84GTHH277 pKa = 6.14KK278 pKa = 10.08LQARR282 pKa = 11.84GTHH285 pKa = 5.3KK286 pKa = 9.95QQARR290 pKa = 11.84GTHH293 pKa = 6.14KK294 pKa = 10.08LQARR298 pKa = 11.84GTHH301 pKa = 5.3KK302 pKa = 9.95QQARR306 pKa = 11.84GTHH309 pKa = 6.14KK310 pKa = 10.08LQARR314 pKa = 11.84GTHH317 pKa = 5.3KK318 pKa = 9.95QQARR322 pKa = 11.84GTHH325 pKa = 6.14KK326 pKa = 10.08LQARR330 pKa = 11.84GTHH333 pKa = 5.3KK334 pKa = 9.95QQARR338 pKa = 11.84GTHH341 pKa = 5.06KK342 pKa = 9.4QQARR346 pKa = 11.84GMHH349 pKa = 5.64KK350 pKa = 8.68QQARR354 pKa = 11.84GTHH357 pKa = 6.08KK358 pKa = 10.1LQARR362 pKa = 11.84GMHH365 pKa = 5.72KK366 pKa = 9.25QQARR370 pKa = 11.84GMHH373 pKa = 5.64KK374 pKa = 8.68QQARR378 pKa = 11.84GTHH381 pKa = 6.14KK382 pKa = 10.08LQARR386 pKa = 11.84GTHH389 pKa = 5.3KK390 pKa = 9.95QQARR394 pKa = 11.84GTHH397 pKa = 5.09KK398 pKa = 9.34QQARR402 pKa = 11.84GTHH405 pKa = 5.09KK406 pKa = 9.34QQARR410 pKa = 11.84GTHH413 pKa = 6.08KK414 pKa = 10.1LQARR418 pKa = 11.84GMHH421 pKa = 5.72KK422 pKa = 9.25QQARR426 pKa = 11.84GMHH429 pKa = 5.59KK430 pKa = 8.75QQARR434 pKa = 11.84GMHH437 pKa = 5.59KK438 pKa = 8.75QQARR442 pKa = 11.84GMHH445 pKa = 5.59KK446 pKa = 8.75QQARR450 pKa = 11.84GMHH453 pKa = 5.59KK454 pKa = 8.75QQARR458 pKa = 11.84GMHH461 pKa = 5.66KK462 pKa = 9.9QQANLTAVPIHH473 pKa = 6.65CNNMRR478 pKa = 11.84HH479 pKa = 6.44CII481 pKa = 3.75
MM1 pKa = 7.31TGYY4 pKa = 7.86TAAAVSVTPGKK15 pKa = 10.07CCRR18 pKa = 11.84GVHH21 pKa = 5.99KK22 pKa = 10.61LQARR26 pKa = 11.84GMHH29 pKa = 5.72KK30 pKa = 9.25QQARR34 pKa = 11.84GMHH37 pKa = 6.1KK38 pKa = 8.58PQARR42 pKa = 11.84GMHH45 pKa = 6.11KK46 pKa = 8.84PQARR50 pKa = 11.84GMHH53 pKa = 5.6KK54 pKa = 8.96QQARR58 pKa = 11.84GMHH61 pKa = 5.59KK62 pKa = 8.75QQARR66 pKa = 11.84GMHH69 pKa = 6.1KK70 pKa = 8.58PQARR74 pKa = 11.84GMHH77 pKa = 5.6KK78 pKa = 8.96QQARR82 pKa = 11.84GMHH85 pKa = 5.59KK86 pKa = 8.75QQARR90 pKa = 11.84GMHH93 pKa = 5.59KK94 pKa = 8.75QQARR98 pKa = 11.84GMHH101 pKa = 5.59KK102 pKa = 8.75QQARR106 pKa = 11.84GMHH109 pKa = 5.59KK110 pKa = 8.75QQARR114 pKa = 11.84GMHH117 pKa = 6.1KK118 pKa = 8.58PQARR122 pKa = 11.84GMHH125 pKa = 5.6KK126 pKa = 8.96QQARR130 pKa = 11.84GMHH133 pKa = 6.1KK134 pKa = 8.58PQARR138 pKa = 11.84GMHH141 pKa = 5.6KK142 pKa = 8.96QQARR146 pKa = 11.84GMHH149 pKa = 5.59KK150 pKa = 8.75QQARR154 pKa = 11.84GMHH157 pKa = 5.59KK158 pKa = 8.75QQARR162 pKa = 11.84GMHH165 pKa = 5.64KK166 pKa = 8.68QQARR170 pKa = 11.84GTHH173 pKa = 5.09KK174 pKa = 9.34QQARR178 pKa = 11.84GTHH181 pKa = 5.09KK182 pKa = 9.34QQARR186 pKa = 11.84GTHH189 pKa = 5.09KK190 pKa = 9.34QQARR194 pKa = 11.84GTHH197 pKa = 5.09KK198 pKa = 9.34QQARR202 pKa = 11.84GTHH205 pKa = 5.09KK206 pKa = 9.34QQARR210 pKa = 11.84GTHH213 pKa = 5.09KK214 pKa = 9.34QQARR218 pKa = 11.84GTHH221 pKa = 5.09KK222 pKa = 9.34QQARR226 pKa = 11.84GTHH229 pKa = 5.09KK230 pKa = 9.34QQARR234 pKa = 11.84GTHH237 pKa = 6.14KK238 pKa = 10.08LQARR242 pKa = 11.84GTHH245 pKa = 6.34KK246 pKa = 10.43LQARR250 pKa = 11.84GTHH253 pKa = 5.3KK254 pKa = 9.95QQARR258 pKa = 11.84GTHH261 pKa = 6.14KK262 pKa = 10.08LQARR266 pKa = 11.84GTHH269 pKa = 5.3KK270 pKa = 9.95QQARR274 pKa = 11.84GTHH277 pKa = 6.14KK278 pKa = 10.08LQARR282 pKa = 11.84GTHH285 pKa = 5.3KK286 pKa = 9.95QQARR290 pKa = 11.84GTHH293 pKa = 6.14KK294 pKa = 10.08LQARR298 pKa = 11.84GTHH301 pKa = 5.3KK302 pKa = 9.95QQARR306 pKa = 11.84GTHH309 pKa = 6.14KK310 pKa = 10.08LQARR314 pKa = 11.84GTHH317 pKa = 5.3KK318 pKa = 9.95QQARR322 pKa = 11.84GTHH325 pKa = 6.14KK326 pKa = 10.08LQARR330 pKa = 11.84GTHH333 pKa = 5.3KK334 pKa = 9.95QQARR338 pKa = 11.84GTHH341 pKa = 5.06KK342 pKa = 9.4QQARR346 pKa = 11.84GMHH349 pKa = 5.64KK350 pKa = 8.68QQARR354 pKa = 11.84GTHH357 pKa = 6.08KK358 pKa = 10.1LQARR362 pKa = 11.84GMHH365 pKa = 5.72KK366 pKa = 9.25QQARR370 pKa = 11.84GMHH373 pKa = 5.64KK374 pKa = 8.68QQARR378 pKa = 11.84GTHH381 pKa = 6.14KK382 pKa = 10.08LQARR386 pKa = 11.84GTHH389 pKa = 5.3KK390 pKa = 9.95QQARR394 pKa = 11.84GTHH397 pKa = 5.09KK398 pKa = 9.34QQARR402 pKa = 11.84GTHH405 pKa = 5.09KK406 pKa = 9.34QQARR410 pKa = 11.84GTHH413 pKa = 6.08KK414 pKa = 10.1LQARR418 pKa = 11.84GMHH421 pKa = 5.72KK422 pKa = 9.25QQARR426 pKa = 11.84GMHH429 pKa = 5.59KK430 pKa = 8.75QQARR434 pKa = 11.84GMHH437 pKa = 5.59KK438 pKa = 8.75QQARR442 pKa = 11.84GMHH445 pKa = 5.59KK446 pKa = 8.75QQARR450 pKa = 11.84GMHH453 pKa = 5.59KK454 pKa = 8.75QQARR458 pKa = 11.84GMHH461 pKa = 5.66KK462 pKa = 9.9QQANLTAVPIHH473 pKa = 6.65CNNMRR478 pKa = 11.84HH479 pKa = 6.44CII481 pKa = 3.75
Molecular weight: 54.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
29680869 |
18 |
10837 |
640.9 |
71.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.096 ± 0.01 | 2.308 ± 0.011 |
5.08 ± 0.008 | 7.052 ± 0.016 |
3.685 ± 0.008 | 5.917 ± 0.017 |
2.599 ± 0.008 | 5.103 ± 0.01 |
6.34 ± 0.016 | 9.382 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.274 ± 0.005 | 4.337 ± 0.007 |
5.495 ± 0.016 | 4.722 ± 0.011 |
5.175 ± 0.012 | 8.579 ± 0.018 |
5.748 ± 0.022 | 6.028 ± 0.01 |
1.145 ± 0.004 | 2.935 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
