
Nipah virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Henipavirus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
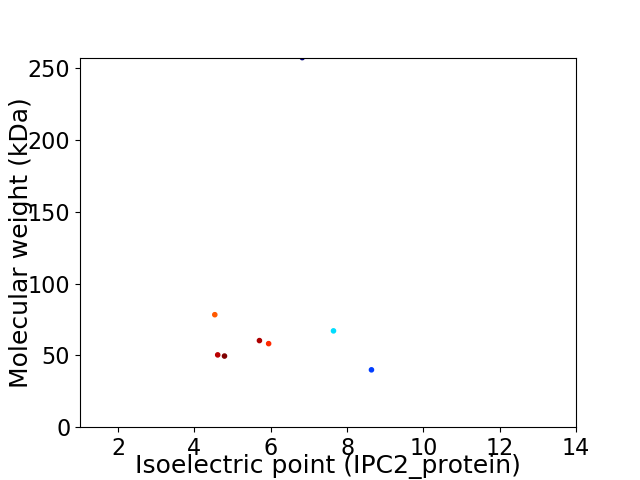
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9IK92|NCAP_NIPAV Nucleoprotein OS=Nipah virus OX=121791 GN=N PE=1 SV=1
MM1 pKa = 8.19DD2 pKa = 5.21KK3 pKa = 11.41LEE5 pKa = 4.44LVNDD9 pKa = 4.35GLNIIDD15 pKa = 5.98FIQKK19 pKa = 7.95NQKK22 pKa = 9.6EE23 pKa = 4.2IQKK26 pKa = 9.07TYY28 pKa = 10.35GRR30 pKa = 11.84SSIQQPSIKK39 pKa = 10.19DD40 pKa = 3.28QTKK43 pKa = 8.84AWEE46 pKa = 4.83DD47 pKa = 3.53FLQCTSGEE55 pKa = 4.26SEE57 pKa = 4.09QVEE60 pKa = 4.25GGMSKK65 pKa = 10.96DD66 pKa = 3.5DD67 pKa = 3.67GDD69 pKa = 3.75VEE71 pKa = 4.45RR72 pKa = 11.84RR73 pKa = 11.84NLEE76 pKa = 4.35DD77 pKa = 4.48LSSTSPTDD85 pKa = 3.19GTIGKK90 pKa = 9.18RR91 pKa = 11.84VSNTRR96 pKa = 11.84DD97 pKa = 2.99WAEE100 pKa = 3.94GSDD103 pKa = 5.29DD104 pKa = 3.66IQLDD108 pKa = 3.81PVVTDD113 pKa = 3.48VVYY116 pKa = 10.5HH117 pKa = 5.48DD118 pKa = 5.08HH119 pKa = 7.06GGEE122 pKa = 4.01CTGYY126 pKa = 10.79GFTSSPEE133 pKa = 4.18RR134 pKa = 11.84GWSDD138 pKa = 3.45YY139 pKa = 11.32TSGANNGNVCLVSDD153 pKa = 4.63AKK155 pKa = 9.89MLSYY159 pKa = 10.77APEE162 pKa = 3.89IAVSKK167 pKa = 10.4EE168 pKa = 3.5DD169 pKa = 3.68RR170 pKa = 11.84EE171 pKa = 4.16TDD173 pKa = 4.02LVHH176 pKa = 7.22LEE178 pKa = 4.42NKK180 pKa = 10.43LSTTGLNPTAVPFTLRR196 pKa = 11.84NLSDD200 pKa = 3.72PAKK203 pKa = 9.53DD204 pKa = 3.45SPVIAEE210 pKa = 4.48HH211 pKa = 6.52YY212 pKa = 9.65YY213 pKa = 10.22GLGVKK218 pKa = 8.05EE219 pKa = 4.15QNVGPQTSRR228 pKa = 11.84NVNLDD233 pKa = 3.46SIKK236 pKa = 10.65LYY238 pKa = 10.66TSDD241 pKa = 5.95DD242 pKa = 3.83EE243 pKa = 5.1EE244 pKa = 6.27ADD246 pKa = 3.34QLEE249 pKa = 4.58FEE251 pKa = 5.23DD252 pKa = 4.72EE253 pKa = 4.41FAGSSSEE260 pKa = 4.27VIVGISPEE268 pKa = 4.08DD269 pKa = 3.79EE270 pKa = 4.33EE271 pKa = 4.58PSSVGGKK278 pKa = 9.12PNEE281 pKa = 4.58SIGRR285 pKa = 11.84TIEE288 pKa = 4.16GQSIRR293 pKa = 11.84DD294 pKa = 3.52NLQAKK299 pKa = 9.31DD300 pKa = 3.86NKK302 pKa = 9.73STDD305 pKa = 3.41VPGAGPKK312 pKa = 10.25DD313 pKa = 3.52SAVKK317 pKa = 10.37EE318 pKa = 4.11EE319 pKa = 4.44PPQKK323 pKa = 9.86RR324 pKa = 11.84LPMLAEE330 pKa = 4.08EE331 pKa = 5.05FEE333 pKa = 4.85CSGSEE338 pKa = 3.96DD339 pKa = 4.05PIIRR343 pKa = 11.84EE344 pKa = 3.95LLKK347 pKa = 10.89EE348 pKa = 3.86NSLINCQQGKK358 pKa = 9.45DD359 pKa = 3.68AQPPYY364 pKa = 8.9HH365 pKa = 6.68WSIEE369 pKa = 4.04RR370 pKa = 11.84SISPDD375 pKa = 2.53KK376 pKa = 11.25TEE378 pKa = 3.96IVNGAVQTADD388 pKa = 3.47RR389 pKa = 11.84QRR391 pKa = 11.84PGTPMPKK398 pKa = 9.87SRR400 pKa = 11.84GIPIKK405 pKa = 10.52KK406 pKa = 8.66GTDD409 pKa = 2.9AKK411 pKa = 10.69YY412 pKa = 10.28PSAGTEE418 pKa = 3.81NVPGSKK424 pKa = 9.92SGATRR429 pKa = 11.84HH430 pKa = 5.28VRR432 pKa = 11.84GSPPYY437 pKa = 10.27QEE439 pKa = 4.47GKK441 pKa = 10.06SVNAEE446 pKa = 3.78NVQLNASTAVKK457 pKa = 8.92EE458 pKa = 4.36TDD460 pKa = 3.12KK461 pKa = 11.81SEE463 pKa = 4.05VNPVDD468 pKa = 5.69DD469 pKa = 5.81NDD471 pKa = 4.34SLDD474 pKa = 4.4DD475 pKa = 4.26KK476 pKa = 11.55YY477 pKa = 11.65IMPSDD482 pKa = 4.05DD483 pKa = 3.52FSNTFFPHH491 pKa = 5.87DD492 pKa = 3.84TDD494 pKa = 3.38RR495 pKa = 11.84LNYY498 pKa = 9.73HH499 pKa = 7.03ADD501 pKa = 3.2HH502 pKa = 6.79LGDD505 pKa = 3.89YY506 pKa = 10.71DD507 pKa = 6.17LEE509 pKa = 4.55TLCEE513 pKa = 4.02EE514 pKa = 4.32SVLMGVINSIKK525 pKa = 10.39LINLDD530 pKa = 3.0MRR532 pKa = 11.84LNHH535 pKa = 6.87IEE537 pKa = 4.16EE538 pKa = 4.25QVKK541 pKa = 9.97EE542 pKa = 3.88IPKK545 pKa = 9.97IINKK549 pKa = 9.78LEE551 pKa = 4.5SIDD554 pKa = 4.02RR555 pKa = 11.84VLAKK559 pKa = 10.14TNTALSTIEE568 pKa = 3.82GHH570 pKa = 6.27LVSMMIMIPGKK581 pKa = 10.71GKK583 pKa = 10.48GEE585 pKa = 3.69RR586 pKa = 11.84KK587 pKa = 9.75GKK589 pKa = 10.07NNPEE593 pKa = 4.17LKK595 pKa = 10.22PVIGRR600 pKa = 11.84DD601 pKa = 3.04ILEE604 pKa = 4.16QQSLFSFDD612 pKa = 3.07NVKK615 pKa = 10.48NFRR618 pKa = 11.84DD619 pKa = 3.83GSLTNEE625 pKa = 4.21PYY627 pKa = 10.58GAAVQLRR634 pKa = 11.84EE635 pKa = 4.25DD636 pKa = 4.91LILPEE641 pKa = 4.92LNFEE645 pKa = 4.33EE646 pKa = 4.79TNASQFVPMADD657 pKa = 3.3DD658 pKa = 3.53SSRR661 pKa = 11.84DD662 pKa = 3.63VIKK665 pKa = 9.91TLIRR669 pKa = 11.84THH671 pKa = 5.98IKK673 pKa = 10.44DD674 pKa = 3.3RR675 pKa = 11.84EE676 pKa = 4.2LRR678 pKa = 11.84SEE680 pKa = 4.69LIGYY684 pKa = 7.78LNKK687 pKa = 10.56AEE689 pKa = 4.19NDD691 pKa = 3.48EE692 pKa = 4.69EE693 pKa = 4.13IQEE696 pKa = 4.22IANTVNDD703 pKa = 4.81IIDD706 pKa = 3.68GNII709 pKa = 2.98
MM1 pKa = 8.19DD2 pKa = 5.21KK3 pKa = 11.41LEE5 pKa = 4.44LVNDD9 pKa = 4.35GLNIIDD15 pKa = 5.98FIQKK19 pKa = 7.95NQKK22 pKa = 9.6EE23 pKa = 4.2IQKK26 pKa = 9.07TYY28 pKa = 10.35GRR30 pKa = 11.84SSIQQPSIKK39 pKa = 10.19DD40 pKa = 3.28QTKK43 pKa = 8.84AWEE46 pKa = 4.83DD47 pKa = 3.53FLQCTSGEE55 pKa = 4.26SEE57 pKa = 4.09QVEE60 pKa = 4.25GGMSKK65 pKa = 10.96DD66 pKa = 3.5DD67 pKa = 3.67GDD69 pKa = 3.75VEE71 pKa = 4.45RR72 pKa = 11.84RR73 pKa = 11.84NLEE76 pKa = 4.35DD77 pKa = 4.48LSSTSPTDD85 pKa = 3.19GTIGKK90 pKa = 9.18RR91 pKa = 11.84VSNTRR96 pKa = 11.84DD97 pKa = 2.99WAEE100 pKa = 3.94GSDD103 pKa = 5.29DD104 pKa = 3.66IQLDD108 pKa = 3.81PVVTDD113 pKa = 3.48VVYY116 pKa = 10.5HH117 pKa = 5.48DD118 pKa = 5.08HH119 pKa = 7.06GGEE122 pKa = 4.01CTGYY126 pKa = 10.79GFTSSPEE133 pKa = 4.18RR134 pKa = 11.84GWSDD138 pKa = 3.45YY139 pKa = 11.32TSGANNGNVCLVSDD153 pKa = 4.63AKK155 pKa = 9.89MLSYY159 pKa = 10.77APEE162 pKa = 3.89IAVSKK167 pKa = 10.4EE168 pKa = 3.5DD169 pKa = 3.68RR170 pKa = 11.84EE171 pKa = 4.16TDD173 pKa = 4.02LVHH176 pKa = 7.22LEE178 pKa = 4.42NKK180 pKa = 10.43LSTTGLNPTAVPFTLRR196 pKa = 11.84NLSDD200 pKa = 3.72PAKK203 pKa = 9.53DD204 pKa = 3.45SPVIAEE210 pKa = 4.48HH211 pKa = 6.52YY212 pKa = 9.65YY213 pKa = 10.22GLGVKK218 pKa = 8.05EE219 pKa = 4.15QNVGPQTSRR228 pKa = 11.84NVNLDD233 pKa = 3.46SIKK236 pKa = 10.65LYY238 pKa = 10.66TSDD241 pKa = 5.95DD242 pKa = 3.83EE243 pKa = 5.1EE244 pKa = 6.27ADD246 pKa = 3.34QLEE249 pKa = 4.58FEE251 pKa = 5.23DD252 pKa = 4.72EE253 pKa = 4.41FAGSSSEE260 pKa = 4.27VIVGISPEE268 pKa = 4.08DD269 pKa = 3.79EE270 pKa = 4.33EE271 pKa = 4.58PSSVGGKK278 pKa = 9.12PNEE281 pKa = 4.58SIGRR285 pKa = 11.84TIEE288 pKa = 4.16GQSIRR293 pKa = 11.84DD294 pKa = 3.52NLQAKK299 pKa = 9.31DD300 pKa = 3.86NKK302 pKa = 9.73STDD305 pKa = 3.41VPGAGPKK312 pKa = 10.25DD313 pKa = 3.52SAVKK317 pKa = 10.37EE318 pKa = 4.11EE319 pKa = 4.44PPQKK323 pKa = 9.86RR324 pKa = 11.84LPMLAEE330 pKa = 4.08EE331 pKa = 5.05FEE333 pKa = 4.85CSGSEE338 pKa = 3.96DD339 pKa = 4.05PIIRR343 pKa = 11.84EE344 pKa = 3.95LLKK347 pKa = 10.89EE348 pKa = 3.86NSLINCQQGKK358 pKa = 9.45DD359 pKa = 3.68AQPPYY364 pKa = 8.9HH365 pKa = 6.68WSIEE369 pKa = 4.04RR370 pKa = 11.84SISPDD375 pKa = 2.53KK376 pKa = 11.25TEE378 pKa = 3.96IVNGAVQTADD388 pKa = 3.47RR389 pKa = 11.84QRR391 pKa = 11.84PGTPMPKK398 pKa = 9.87SRR400 pKa = 11.84GIPIKK405 pKa = 10.52KK406 pKa = 8.66GTDD409 pKa = 2.9AKK411 pKa = 10.69YY412 pKa = 10.28PSAGTEE418 pKa = 3.81NVPGSKK424 pKa = 9.92SGATRR429 pKa = 11.84HH430 pKa = 5.28VRR432 pKa = 11.84GSPPYY437 pKa = 10.27QEE439 pKa = 4.47GKK441 pKa = 10.06SVNAEE446 pKa = 3.78NVQLNASTAVKK457 pKa = 8.92EE458 pKa = 4.36TDD460 pKa = 3.12KK461 pKa = 11.81SEE463 pKa = 4.05VNPVDD468 pKa = 5.69DD469 pKa = 5.81NDD471 pKa = 4.34SLDD474 pKa = 4.4DD475 pKa = 4.26KK476 pKa = 11.55YY477 pKa = 11.65IMPSDD482 pKa = 4.05DD483 pKa = 3.52FSNTFFPHH491 pKa = 5.87DD492 pKa = 3.84TDD494 pKa = 3.38RR495 pKa = 11.84LNYY498 pKa = 9.73HH499 pKa = 7.03ADD501 pKa = 3.2HH502 pKa = 6.79LGDD505 pKa = 3.89YY506 pKa = 10.71DD507 pKa = 6.17LEE509 pKa = 4.55TLCEE513 pKa = 4.02EE514 pKa = 4.32SVLMGVINSIKK525 pKa = 10.39LINLDD530 pKa = 3.0MRR532 pKa = 11.84LNHH535 pKa = 6.87IEE537 pKa = 4.16EE538 pKa = 4.25QVKK541 pKa = 9.97EE542 pKa = 3.88IPKK545 pKa = 9.97IINKK549 pKa = 9.78LEE551 pKa = 4.5SIDD554 pKa = 4.02RR555 pKa = 11.84VLAKK559 pKa = 10.14TNTALSTIEE568 pKa = 3.82GHH570 pKa = 6.27LVSMMIMIPGKK581 pKa = 10.71GKK583 pKa = 10.48GEE585 pKa = 3.69RR586 pKa = 11.84KK587 pKa = 9.75GKK589 pKa = 10.07NNPEE593 pKa = 4.17LKK595 pKa = 10.22PVIGRR600 pKa = 11.84DD601 pKa = 3.04ILEE604 pKa = 4.16QQSLFSFDD612 pKa = 3.07NVKK615 pKa = 10.48NFRR618 pKa = 11.84DD619 pKa = 3.83GSLTNEE625 pKa = 4.21PYY627 pKa = 10.58GAAVQLRR634 pKa = 11.84EE635 pKa = 4.25DD636 pKa = 4.91LILPEE641 pKa = 4.92LNFEE645 pKa = 4.33EE646 pKa = 4.79TNASQFVPMADD657 pKa = 3.3DD658 pKa = 3.53SSRR661 pKa = 11.84DD662 pKa = 3.63VIKK665 pKa = 9.91TLIRR669 pKa = 11.84THH671 pKa = 5.98IKK673 pKa = 10.44DD674 pKa = 3.3RR675 pKa = 11.84EE676 pKa = 4.2LRR678 pKa = 11.84SEE680 pKa = 4.69LIGYY684 pKa = 7.78LNKK687 pKa = 10.56AEE689 pKa = 4.19NDD691 pKa = 3.48EE692 pKa = 4.69EE693 pKa = 4.13IQEE696 pKa = 4.22IANTVNDD703 pKa = 4.81IIDD706 pKa = 3.68GNII709 pKa = 2.98
Molecular weight: 78.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9IK91|PHOSP_NIPAV Phosphoprotein OS=Nipah virus OX=121791 GN=P/V/C PE=1 SV=1
MM1 pKa = 7.51EE2 pKa = 6.3PDD4 pKa = 3.13IKK6 pKa = 10.9SISSEE11 pKa = 3.97SMEE14 pKa = 4.14GVSDD18 pKa = 4.46FSPSSWEE25 pKa = 3.6HH26 pKa = 5.58GGYY29 pKa = 9.7LDD31 pKa = 4.68KK32 pKa = 11.44VEE34 pKa = 4.88PEE36 pKa = 3.47IDD38 pKa = 3.8EE39 pKa = 4.53NGSMIPKK46 pKa = 9.67YY47 pKa = 10.46KK48 pKa = 10.14IYY50 pKa = 10.47TPGANEE56 pKa = 3.76RR57 pKa = 11.84KK58 pKa = 9.16YY59 pKa = 11.08NNYY62 pKa = 9.42MYY64 pKa = 9.67LICYY68 pKa = 8.93GFVEE72 pKa = 4.58DD73 pKa = 5.22VEE75 pKa = 4.54RR76 pKa = 11.84TPEE79 pKa = 3.53TGKK82 pKa = 10.18RR83 pKa = 11.84KK84 pKa = 9.95KK85 pKa = 9.99IRR87 pKa = 11.84TIAAYY92 pKa = 8.88PLGVGKK98 pKa = 10.17SASHH102 pKa = 6.01PQDD105 pKa = 3.81LLEE108 pKa = 4.51EE109 pKa = 4.4LCSLKK114 pKa = 9.7VTVRR118 pKa = 11.84RR119 pKa = 11.84TAGSTEE125 pKa = 3.97KK126 pKa = 10.43IVFGSSGPLNHH137 pKa = 6.8LVPWKK142 pKa = 10.41KK143 pKa = 10.57VLTSGSIFNAVKK155 pKa = 9.82VCRR158 pKa = 11.84NVDD161 pKa = 3.48QIQLDD166 pKa = 3.4KK167 pKa = 11.11HH168 pKa = 4.46QALRR172 pKa = 11.84IFFLSITKK180 pKa = 10.61LNDD183 pKa = 2.92SGIYY187 pKa = 8.39MIPRR191 pKa = 11.84TMLEE195 pKa = 3.67FRR197 pKa = 11.84RR198 pKa = 11.84NNAIAFNLLVYY209 pKa = 10.11LKK211 pKa = 10.14IDD213 pKa = 3.74ADD215 pKa = 3.98LSKK218 pKa = 10.54MGIQGSLDD226 pKa = 3.2KK227 pKa = 11.43DD228 pKa = 3.83GFKK231 pKa = 10.66VASFMLHH238 pKa = 6.3LGNFVRR244 pKa = 11.84RR245 pKa = 11.84AGKK248 pKa = 10.17YY249 pKa = 9.66YY250 pKa = 10.69SVDD253 pKa = 3.09YY254 pKa = 10.53CRR256 pKa = 11.84RR257 pKa = 11.84KK258 pKa = 8.96IDD260 pKa = 3.4RR261 pKa = 11.84MKK263 pKa = 10.9LQFSLGSIGGLSLHH277 pKa = 6.47IKK279 pKa = 9.92INGVISKK286 pKa = 10.48RR287 pKa = 11.84LFAQMGFQKK296 pKa = 10.44NLCFSLMDD304 pKa = 3.9INPWLNRR311 pKa = 11.84LTWNNSCEE319 pKa = 4.12ISRR322 pKa = 11.84VAAVLQPSIPRR333 pKa = 11.84EE334 pKa = 3.57FMIYY338 pKa = 10.7DD339 pKa = 3.94DD340 pKa = 4.73VFIDD344 pKa = 3.35NTGRR348 pKa = 11.84ILKK351 pKa = 9.94GG352 pKa = 3.12
MM1 pKa = 7.51EE2 pKa = 6.3PDD4 pKa = 3.13IKK6 pKa = 10.9SISSEE11 pKa = 3.97SMEE14 pKa = 4.14GVSDD18 pKa = 4.46FSPSSWEE25 pKa = 3.6HH26 pKa = 5.58GGYY29 pKa = 9.7LDD31 pKa = 4.68KK32 pKa = 11.44VEE34 pKa = 4.88PEE36 pKa = 3.47IDD38 pKa = 3.8EE39 pKa = 4.53NGSMIPKK46 pKa = 9.67YY47 pKa = 10.46KK48 pKa = 10.14IYY50 pKa = 10.47TPGANEE56 pKa = 3.76RR57 pKa = 11.84KK58 pKa = 9.16YY59 pKa = 11.08NNYY62 pKa = 9.42MYY64 pKa = 9.67LICYY68 pKa = 8.93GFVEE72 pKa = 4.58DD73 pKa = 5.22VEE75 pKa = 4.54RR76 pKa = 11.84TPEE79 pKa = 3.53TGKK82 pKa = 10.18RR83 pKa = 11.84KK84 pKa = 9.95KK85 pKa = 9.99IRR87 pKa = 11.84TIAAYY92 pKa = 8.88PLGVGKK98 pKa = 10.17SASHH102 pKa = 6.01PQDD105 pKa = 3.81LLEE108 pKa = 4.51EE109 pKa = 4.4LCSLKK114 pKa = 9.7VTVRR118 pKa = 11.84RR119 pKa = 11.84TAGSTEE125 pKa = 3.97KK126 pKa = 10.43IVFGSSGPLNHH137 pKa = 6.8LVPWKK142 pKa = 10.41KK143 pKa = 10.57VLTSGSIFNAVKK155 pKa = 9.82VCRR158 pKa = 11.84NVDD161 pKa = 3.48QIQLDD166 pKa = 3.4KK167 pKa = 11.11HH168 pKa = 4.46QALRR172 pKa = 11.84IFFLSITKK180 pKa = 10.61LNDD183 pKa = 2.92SGIYY187 pKa = 8.39MIPRR191 pKa = 11.84TMLEE195 pKa = 3.67FRR197 pKa = 11.84RR198 pKa = 11.84NNAIAFNLLVYY209 pKa = 10.11LKK211 pKa = 10.14IDD213 pKa = 3.74ADD215 pKa = 3.98LSKK218 pKa = 10.54MGIQGSLDD226 pKa = 3.2KK227 pKa = 11.43DD228 pKa = 3.83GFKK231 pKa = 10.66VASFMLHH238 pKa = 6.3LGNFVRR244 pKa = 11.84RR245 pKa = 11.84AGKK248 pKa = 10.17YY249 pKa = 9.66YY250 pKa = 10.69SVDD253 pKa = 3.09YY254 pKa = 10.53CRR256 pKa = 11.84RR257 pKa = 11.84KK258 pKa = 8.96IDD260 pKa = 3.4RR261 pKa = 11.84MKK263 pKa = 10.9LQFSLGSIGGLSLHH277 pKa = 6.47IKK279 pKa = 9.92INGVISKK286 pKa = 10.48RR287 pKa = 11.84LFAQMGFQKK296 pKa = 10.44NLCFSLMDD304 pKa = 3.9INPWLNRR311 pKa = 11.84LTWNNSCEE319 pKa = 4.12ISRR322 pKa = 11.84VAAVLQPSIPRR333 pKa = 11.84EE334 pKa = 3.57FMIYY338 pKa = 10.7DD339 pKa = 3.94DD340 pKa = 4.73VFIDD344 pKa = 3.35NTGRR348 pKa = 11.84ILKK351 pKa = 9.94GG352 pKa = 3.12
Molecular weight: 39.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5891 |
352 |
2244 |
736.4 |
82.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.025 ± 0.637 | 1.731 ± 0.274 |
6.281 ± 0.535 | 6.569 ± 0.644 |
3.191 ± 0.369 | 6.043 ± 0.699 |
1.765 ± 0.381 | 8.046 ± 0.559 |
6.315 ± 0.268 | 8.98 ± 0.77 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.217 | 5.805 ± 0.316 |
4.651 ± 0.474 | 3.768 ± 0.263 |
5.008 ± 0.324 | 8.861 ± 0.221 |
5.483 ± 0.253 | 5.839 ± 0.277 |
0.985 ± 0.131 | 3.327 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
