
Thrips-associated genomovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus echi1
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
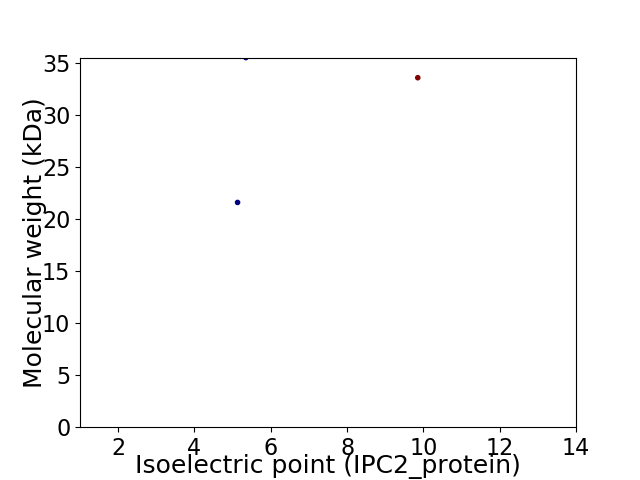
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
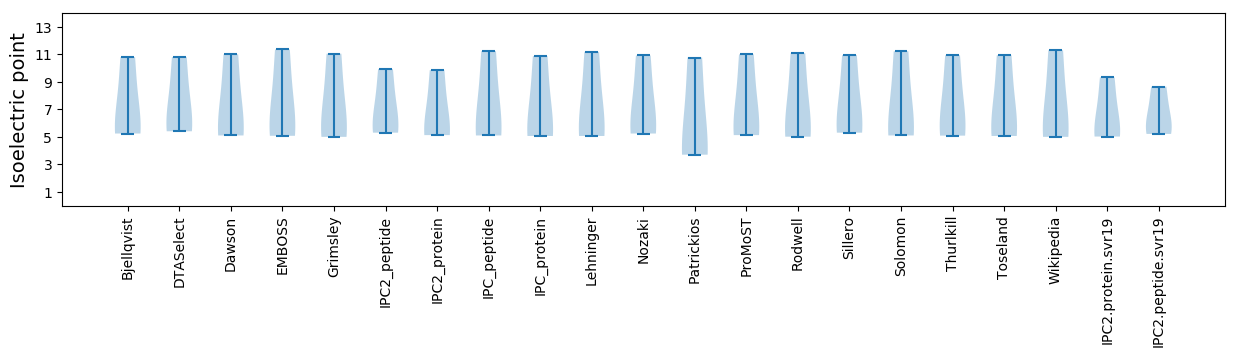
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8YT72|A0A1P8YT72_9VIRU RepA OS=Thrips-associated genomovirus 3 OX=1941237 PE=3 SV=1
MM1 pKa = 7.97PFRR4 pKa = 11.84FAAKK8 pKa = 10.01YY9 pKa = 10.66GLLTYY14 pKa = 7.2PQCGDD19 pKa = 3.72LDD21 pKa = 3.7PWSVSDD27 pKa = 3.44HH28 pKa = 6.45LGRR31 pKa = 11.84LGAEE35 pKa = 4.22CIIGRR40 pKa = 11.84EE41 pKa = 4.15DD42 pKa = 3.37HH43 pKa = 7.1SDD45 pKa = 3.54GGVHH49 pKa = 6.1LHH51 pKa = 6.35AFFMFEE57 pKa = 3.8RR58 pKa = 11.84RR59 pKa = 11.84FEE61 pKa = 4.22SRR63 pKa = 11.84DD64 pKa = 3.13VRR66 pKa = 11.84VFDD69 pKa = 4.26VEE71 pKa = 4.3GRR73 pKa = 11.84HH74 pKa = 5.7PNVVRR79 pKa = 11.84GYY81 pKa = 7.64STPSKK86 pKa = 10.33GYY88 pKa = 10.77AYY90 pKa = 8.25ATKK93 pKa = 10.63DD94 pKa = 3.16GDD96 pKa = 4.0VVAGGLEE103 pKa = 4.13CPIDD107 pKa = 3.88GASVSEE113 pKa = 5.0ASSKK117 pKa = 7.87WARR120 pKa = 11.84AILAEE125 pKa = 4.02SRR127 pKa = 11.84EE128 pKa = 4.04EE129 pKa = 3.76FFAIVAEE136 pKa = 4.23LDD138 pKa = 3.53PRR140 pKa = 11.84ALCCSFGSLRR150 pKa = 11.84AYY152 pKa = 10.47ADD154 pKa = 2.98WKK156 pKa = 9.94YY157 pKa = 11.07RR158 pKa = 11.84PVLDD162 pKa = 4.7QYY164 pKa = 8.68VTPAGISFCTDD175 pKa = 3.18EE176 pKa = 5.02VPQLHH181 pKa = 7.17DD182 pKa = 3.09WVQRR186 pKa = 11.84NLAGSVVGEE195 pKa = 4.12
MM1 pKa = 7.97PFRR4 pKa = 11.84FAAKK8 pKa = 10.01YY9 pKa = 10.66GLLTYY14 pKa = 7.2PQCGDD19 pKa = 3.72LDD21 pKa = 3.7PWSVSDD27 pKa = 3.44HH28 pKa = 6.45LGRR31 pKa = 11.84LGAEE35 pKa = 4.22CIIGRR40 pKa = 11.84EE41 pKa = 4.15DD42 pKa = 3.37HH43 pKa = 7.1SDD45 pKa = 3.54GGVHH49 pKa = 6.1LHH51 pKa = 6.35AFFMFEE57 pKa = 3.8RR58 pKa = 11.84RR59 pKa = 11.84FEE61 pKa = 4.22SRR63 pKa = 11.84DD64 pKa = 3.13VRR66 pKa = 11.84VFDD69 pKa = 4.26VEE71 pKa = 4.3GRR73 pKa = 11.84HH74 pKa = 5.7PNVVRR79 pKa = 11.84GYY81 pKa = 7.64STPSKK86 pKa = 10.33GYY88 pKa = 10.77AYY90 pKa = 8.25ATKK93 pKa = 10.63DD94 pKa = 3.16GDD96 pKa = 4.0VVAGGLEE103 pKa = 4.13CPIDD107 pKa = 3.88GASVSEE113 pKa = 5.0ASSKK117 pKa = 7.87WARR120 pKa = 11.84AILAEE125 pKa = 4.02SRR127 pKa = 11.84EE128 pKa = 4.04EE129 pKa = 3.76FFAIVAEE136 pKa = 4.23LDD138 pKa = 3.53PRR140 pKa = 11.84ALCCSFGSLRR150 pKa = 11.84AYY152 pKa = 10.47ADD154 pKa = 2.98WKK156 pKa = 9.94YY157 pKa = 11.07RR158 pKa = 11.84PVLDD162 pKa = 4.7QYY164 pKa = 8.68VTPAGISFCTDD175 pKa = 3.18EE176 pKa = 5.02VPQLHH181 pKa = 7.17DD182 pKa = 3.09WVQRR186 pKa = 11.84NLAGSVVGEE195 pKa = 4.12
Molecular weight: 21.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YT72|A0A1P8YT72_9VIRU RepA OS=Thrips-associated genomovirus 3 OX=1941237 PE=3 SV=1
MM1 pKa = 7.0YY2 pKa = 10.13RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84PSKK8 pKa = 10.19RR9 pKa = 11.84RR10 pKa = 11.84SGGSRR15 pKa = 11.84RR16 pKa = 11.84TPRR19 pKa = 11.84SSRR22 pKa = 11.84RR23 pKa = 11.84SYY25 pKa = 11.22AKK27 pKa = 8.42KK28 pKa = 8.0TSYY31 pKa = 10.58RR32 pKa = 11.84RR33 pKa = 11.84TRR35 pKa = 11.84TVRR38 pKa = 11.84KK39 pKa = 7.28LTSRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84LLNITSQKK53 pKa = 10.32KK54 pKa = 8.32RR55 pKa = 11.84DD56 pKa = 3.45NMMPVTTDD64 pKa = 2.6WTGQTPIPGPAVLNGAGRR82 pKa = 11.84STILWCATQRR92 pKa = 11.84DD93 pKa = 4.41RR94 pKa = 11.84ASTPYY99 pKa = 10.06IDD101 pKa = 3.8NSEE104 pKa = 4.12AARR107 pKa = 11.84TSKK110 pKa = 10.79SIFLRR115 pKa = 11.84GVRR118 pKa = 11.84EE119 pKa = 3.9FASIRR124 pKa = 11.84TNSPDD129 pKa = 2.51AWRR132 pKa = 11.84WRR134 pKa = 11.84RR135 pKa = 11.84IIFSVKK141 pKa = 10.3GIFVYY146 pKa = 10.74LPLEE150 pKa = 3.84SRR152 pKa = 11.84FAVEE156 pKa = 4.2TSNQGWSRR164 pKa = 11.84AIPDD168 pKa = 3.64YY169 pKa = 11.02SGSASTTARR178 pKa = 11.84NALEE182 pKa = 3.81ALIFAGEE189 pKa = 4.07AGKK192 pKa = 10.27DD193 pKa = 3.12WASVFTAKK201 pKa = 9.94VDD203 pKa = 3.44HH204 pKa = 6.81SRR206 pKa = 11.84ITPLFDD212 pKa = 2.81KK213 pKa = 10.84TRR215 pKa = 11.84ILRR218 pKa = 11.84SGNQQGSYY226 pKa = 10.28FKK228 pKa = 11.06NKK230 pKa = 9.33LWLPVNKK237 pKa = 9.09TLLYY241 pKa = 10.68ADD243 pKa = 4.41EE244 pKa = 4.51EE245 pKa = 4.79SGNNVQDD252 pKa = 3.95GSLSAISRR260 pKa = 11.84QGVGDD265 pKa = 3.53IFIFDD270 pKa = 5.04LFDD273 pKa = 5.17CSTQNSASSLTFEE286 pKa = 5.37PEE288 pKa = 3.28ATMYY292 pKa = 8.49WHH294 pKa = 6.85EE295 pKa = 4.17RR296 pKa = 3.48
MM1 pKa = 7.0YY2 pKa = 10.13RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84PSKK8 pKa = 10.19RR9 pKa = 11.84RR10 pKa = 11.84SGGSRR15 pKa = 11.84RR16 pKa = 11.84TPRR19 pKa = 11.84SSRR22 pKa = 11.84RR23 pKa = 11.84SYY25 pKa = 11.22AKK27 pKa = 8.42KK28 pKa = 8.0TSYY31 pKa = 10.58RR32 pKa = 11.84RR33 pKa = 11.84TRR35 pKa = 11.84TVRR38 pKa = 11.84KK39 pKa = 7.28LTSRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84LLNITSQKK53 pKa = 10.32KK54 pKa = 8.32RR55 pKa = 11.84DD56 pKa = 3.45NMMPVTTDD64 pKa = 2.6WTGQTPIPGPAVLNGAGRR82 pKa = 11.84STILWCATQRR92 pKa = 11.84DD93 pKa = 4.41RR94 pKa = 11.84ASTPYY99 pKa = 10.06IDD101 pKa = 3.8NSEE104 pKa = 4.12AARR107 pKa = 11.84TSKK110 pKa = 10.79SIFLRR115 pKa = 11.84GVRR118 pKa = 11.84EE119 pKa = 3.9FASIRR124 pKa = 11.84TNSPDD129 pKa = 2.51AWRR132 pKa = 11.84WRR134 pKa = 11.84RR135 pKa = 11.84IIFSVKK141 pKa = 10.3GIFVYY146 pKa = 10.74LPLEE150 pKa = 3.84SRR152 pKa = 11.84FAVEE156 pKa = 4.2TSNQGWSRR164 pKa = 11.84AIPDD168 pKa = 3.64YY169 pKa = 11.02SGSASTTARR178 pKa = 11.84NALEE182 pKa = 3.81ALIFAGEE189 pKa = 4.07AGKK192 pKa = 10.27DD193 pKa = 3.12WASVFTAKK201 pKa = 9.94VDD203 pKa = 3.44HH204 pKa = 6.81SRR206 pKa = 11.84ITPLFDD212 pKa = 2.81KK213 pKa = 10.84TRR215 pKa = 11.84ILRR218 pKa = 11.84SGNQQGSYY226 pKa = 10.28FKK228 pKa = 11.06NKK230 pKa = 9.33LWLPVNKK237 pKa = 9.09TLLYY241 pKa = 10.68ADD243 pKa = 4.41EE244 pKa = 4.51EE245 pKa = 4.79SGNNVQDD252 pKa = 3.95GSLSAISRR260 pKa = 11.84QGVGDD265 pKa = 3.53IFIFDD270 pKa = 5.04LFDD273 pKa = 5.17CSTQNSASSLTFEE286 pKa = 5.37PEE288 pKa = 3.28ATMYY292 pKa = 8.49WHH294 pKa = 6.85EE295 pKa = 4.17RR296 pKa = 3.48
Molecular weight: 33.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
809 |
195 |
318 |
269.7 |
30.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.776 ± 0.443 | 2.101 ± 0.64 |
6.799 ± 0.984 | 4.944 ± 0.581 |
5.315 ± 0.45 | 8.158 ± 0.965 |
2.225 ± 0.714 | 4.326 ± 0.484 |
3.832 ± 0.426 | 7.417 ± 0.609 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.236 ± 0.063 | 2.472 ± 0.854 |
4.574 ± 0.161 | 2.596 ± 0.202 |
8.9 ± 1.58 | 8.776 ± 1.483 |
5.068 ± 1.505 | 6.18 ± 0.958 |
2.472 ± 0.126 | 3.832 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
