
Candidatus Micrarchaeum sp. AZ1
Taxonomy: cellular organisms; Archaea; DPANN group; Candidatus Micrarchaeota; Candidatus Micrarchaeum; unclassified Candidatus Micrarchaeum
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
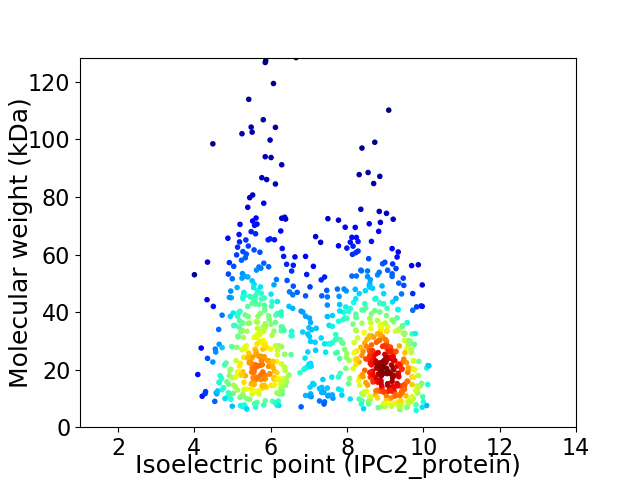
Virtual 2D-PAGE plot for 840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M2YIG1|A0A1M2YIG1_9ARCH Uncharacterized protein OS=Candidatus Micrarchaeum sp. AZ1 OX=1531428 GN=JJ59_01150 PE=4 SV=1
AA1 pKa = 7.36VVPIFFNISNIPTSLQSNPSGIATIDD27 pKa = 3.68GVSYY31 pKa = 10.65SYY33 pKa = 11.38SQLTKK38 pKa = 10.69KK39 pKa = 8.41PTNFDD44 pKa = 3.22WGCNTTHH51 pKa = 7.21TYY53 pKa = 10.78SFDD56 pKa = 4.43PIIYY60 pKa = 9.0NSSGTRR66 pKa = 11.84FVFKK70 pKa = 10.63SVTIDD75 pKa = 3.46GFTSDD80 pKa = 4.5SNSGSATVSTCKK92 pKa = 10.31SQNVTVSYY100 pKa = 10.97AEE102 pKa = 3.87QYY104 pKa = 10.13EE105 pKa = 4.56LYY107 pKa = 10.4DD108 pKa = 3.37IASPSIGGTVSPATSWYY125 pKa = 9.8APSSSVTISEE135 pKa = 4.48TPNTTGHH142 pKa = 5.92YY143 pKa = 10.81VFIDD147 pKa = 3.57WTCTGTGCYY156 pKa = 9.95SGTATSTSVTLNNPINEE173 pKa = 4.27TAVFQSTTTSTSTTSTSTTTSTTSTSTTTSTTSTSTSTTSTSTTTSTSTTTSTSTTSTSTTSTSTTTSTTSTSTSTTSTSTTTSTSTTTSTSTTTSTSTTTSTSTTTSSSTIPFYY288 pKa = 10.0GTCTTNSSSATTFSPSCSNNNGVFSEE314 pKa = 4.71CVAAIYY320 pKa = 10.8QSMSSVDD327 pKa = 3.17WNINSEE333 pKa = 4.19GGGTCQYY340 pKa = 10.72CSTNIGYY347 pKa = 7.61ITNGNACSATADD359 pKa = 3.83SSSSGAIAIGGNPATNYY376 pKa = 9.51QLWATNGSNYY386 pKa = 10.04NSGVQPVFWVNHH398 pKa = 4.42GSIPSSLVILTYY410 pKa = 9.59ATSGGQEE417 pKa = 4.43GILNSTTEE425 pKa = 4.38PGTSAIMYY433 pKa = 9.36DD434 pKa = 3.47GGSFPTYY441 pKa = 9.41GCSVTEE447 pKa = 4.15NLEE450 pKa = 3.74QTTPLFSSTHH460 pKa = 5.44YY461 pKa = 10.51NDD463 pKa = 4.16VYY465 pKa = 11.09ILACGNIPYY474 pKa = 10.02DD475 pKa = 3.99GTGGWIGPEE484 pKa = 4.03LMTGSSTSFSYY495 pKa = 10.69AVYY498 pKa = 10.6VFNPDD503 pKa = 2.48STYY506 pKa = 11.12TFNKK510 pKa = 10.15NSS512 pKa = 3.35
AA1 pKa = 7.36VVPIFFNISNIPTSLQSNPSGIATIDD27 pKa = 3.68GVSYY31 pKa = 10.65SYY33 pKa = 11.38SQLTKK38 pKa = 10.69KK39 pKa = 8.41PTNFDD44 pKa = 3.22WGCNTTHH51 pKa = 7.21TYY53 pKa = 10.78SFDD56 pKa = 4.43PIIYY60 pKa = 9.0NSSGTRR66 pKa = 11.84FVFKK70 pKa = 10.63SVTIDD75 pKa = 3.46GFTSDD80 pKa = 4.5SNSGSATVSTCKK92 pKa = 10.31SQNVTVSYY100 pKa = 10.97AEE102 pKa = 3.87QYY104 pKa = 10.13EE105 pKa = 4.56LYY107 pKa = 10.4DD108 pKa = 3.37IASPSIGGTVSPATSWYY125 pKa = 9.8APSSSVTISEE135 pKa = 4.48TPNTTGHH142 pKa = 5.92YY143 pKa = 10.81VFIDD147 pKa = 3.57WTCTGTGCYY156 pKa = 9.95SGTATSTSVTLNNPINEE173 pKa = 4.27TAVFQSTTTSTSTTSTSTTTSTTSTSTTTSTTSTSTSTTSTSTTTSTSTTTSTSTTSTSTTSTSTTTSTTSTSTSTTSTSTTTSTSTTTSTSTTTSTSTTTSTSTTTSSSTIPFYY288 pKa = 10.0GTCTTNSSSATTFSPSCSNNNGVFSEE314 pKa = 4.71CVAAIYY320 pKa = 10.8QSMSSVDD327 pKa = 3.17WNINSEE333 pKa = 4.19GGGTCQYY340 pKa = 10.72CSTNIGYY347 pKa = 7.61ITNGNACSATADD359 pKa = 3.83SSSSGAIAIGGNPATNYY376 pKa = 9.51QLWATNGSNYY386 pKa = 10.04NSGVQPVFWVNHH398 pKa = 4.42GSIPSSLVILTYY410 pKa = 9.59ATSGGQEE417 pKa = 4.43GILNSTTEE425 pKa = 4.38PGTSAIMYY433 pKa = 9.36DD434 pKa = 3.47GGSFPTYY441 pKa = 9.41GCSVTEE447 pKa = 4.15NLEE450 pKa = 3.74QTTPLFSSTHH460 pKa = 5.44YY461 pKa = 10.51NDD463 pKa = 4.16VYY465 pKa = 11.09ILACGNIPYY474 pKa = 10.02DD475 pKa = 3.99GTGGWIGPEE484 pKa = 4.03LMTGSSTSFSYY495 pKa = 10.69AVYY498 pKa = 10.6VFNPDD503 pKa = 2.48STYY506 pKa = 11.12TFNKK510 pKa = 10.15NSS512 pKa = 3.35
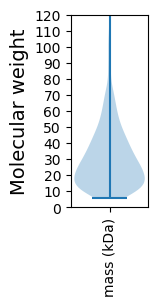
Molecular weight: 52.97 kDa
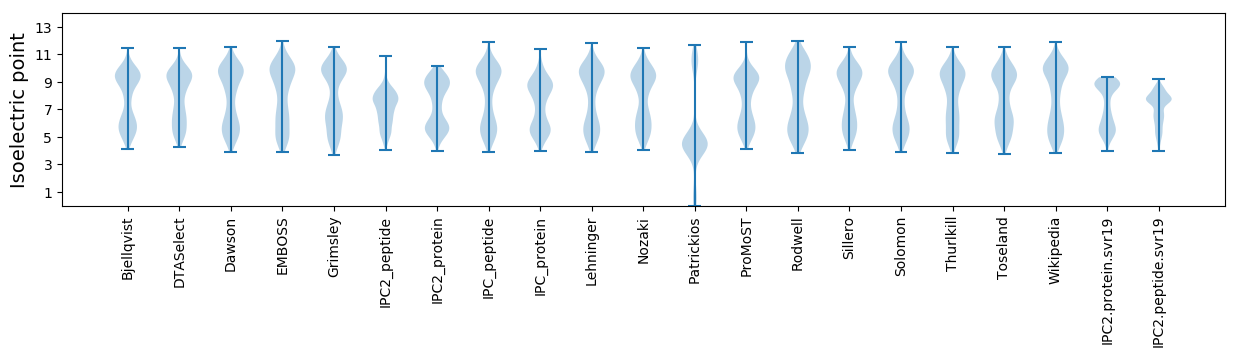
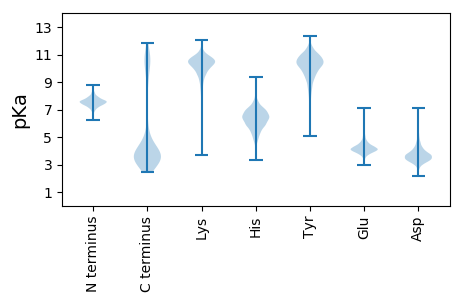
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M2YH48|A0A1M2YH48_9ARCH Uncharacterized protein OS=Candidatus Micrarchaeum sp. AZ1 OX=1531428 GN=JJ59_02780 PE=4 SV=1
MM1 pKa = 7.66PRR3 pKa = 11.84QSPKK7 pKa = 10.4TKK9 pKa = 10.49DD10 pKa = 3.11LARR13 pKa = 11.84AIARR17 pKa = 11.84KK18 pKa = 9.18RR19 pKa = 11.84VAGLISLSEE28 pKa = 3.87KK29 pKa = 10.62ALRR32 pKa = 11.84DD33 pKa = 3.59GNQEE37 pKa = 3.47LSRR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 10.2AEE44 pKa = 4.27LASKK48 pKa = 9.71IQSHH52 pKa = 4.71YY53 pKa = 10.43RR54 pKa = 11.84IRR56 pKa = 11.84PSLRR60 pKa = 11.84YY61 pKa = 8.82RR62 pKa = 11.84VCKK65 pKa = 10.18NCGSILVPGITSSTRR80 pKa = 11.84LSSSRR85 pKa = 11.84GYY87 pKa = 10.18IVVKK91 pKa = 10.51CMVCGTEE98 pKa = 3.63LHH100 pKa = 6.15KK101 pKa = 10.92VYY103 pKa = 10.86KK104 pKa = 10.06KK105 pKa = 10.89
MM1 pKa = 7.66PRR3 pKa = 11.84QSPKK7 pKa = 10.4TKK9 pKa = 10.49DD10 pKa = 3.11LARR13 pKa = 11.84AIARR17 pKa = 11.84KK18 pKa = 9.18RR19 pKa = 11.84VAGLISLSEE28 pKa = 3.87KK29 pKa = 10.62ALRR32 pKa = 11.84DD33 pKa = 3.59GNQEE37 pKa = 3.47LSRR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 10.2AEE44 pKa = 4.27LASKK48 pKa = 9.71IQSHH52 pKa = 4.71YY53 pKa = 10.43RR54 pKa = 11.84IRR56 pKa = 11.84PSLRR60 pKa = 11.84YY61 pKa = 8.82RR62 pKa = 11.84VCKK65 pKa = 10.18NCGSILVPGITSSTRR80 pKa = 11.84LSSSRR85 pKa = 11.84GYY87 pKa = 10.18IVVKK91 pKa = 10.51CMVCGTEE98 pKa = 3.63LHH100 pKa = 6.15KK101 pKa = 10.92VYY103 pKa = 10.86KK104 pKa = 10.06KK105 pKa = 10.89
Molecular weight: 11.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
239489 |
50 |
1152 |
285.1 |
31.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.082 ± 0.086 | 0.703 ± 0.026 |
4.599 ± 0.069 | 6.417 ± 0.121 |
4.292 ± 0.074 | 7.076 ± 0.072 |
1.512 ± 0.03 | 8.288 ± 0.074 |
7.801 ± 0.12 | 8.845 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.016 ± 0.039 | 4.752 ± 0.085 |
3.497 ± 0.052 | 2.365 ± 0.053 |
4.248 ± 0.078 | 8.268 ± 0.115 |
4.831 ± 0.079 | 6.439 ± 0.069 |
0.709 ± 0.024 | 4.261 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
