
Pelagibacter phage HTVC120P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; unclassified Autographiviridae
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
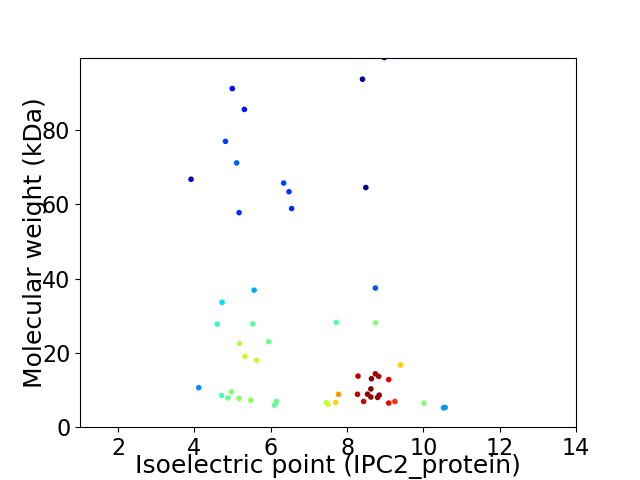
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NSZ1|A0A4Y1NSZ1_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC120P OX=2283021 GN=P120_gp33 PE=4 SV=1
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.29TGNGSTTAYY17 pKa = 9.9SISYY21 pKa = 8.14TYY23 pKa = 10.76RR24 pKa = 11.84DD25 pKa = 3.59AADD28 pKa = 5.02LIVSVNGVATTAYY41 pKa = 9.98SLNAAGTTLTFDD53 pKa = 3.62SAPASSSAIEE63 pKa = 4.0IRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.24TSQTTRR73 pKa = 11.84LTDD76 pKa = 3.47YY77 pKa = 11.08AAGSVLTEE85 pKa = 4.12NDD87 pKa = 3.84LDD89 pKa = 4.08TDD91 pKa = 3.79STQAFFMGQEE101 pKa = 4.68AIDD104 pKa = 4.37DD105 pKa = 4.34ANDD108 pKa = 3.5VIKK111 pKa = 10.73ISSTDD116 pKa = 3.76FQFNAINKK124 pKa = 7.92QIRR127 pKa = 11.84NVANPTSNQDD137 pKa = 3.03VATKK141 pKa = 10.09HH142 pKa = 5.43YY143 pKa = 10.9LEE145 pKa = 4.34NTFLTTANKK154 pKa = 8.32TALTTINANIANIIAVNNNSTNINSAVSNATNINTVATNIGSVNTVAADD203 pKa = 3.22IAKK206 pKa = 10.13VIEE209 pKa = 4.24VANDD213 pKa = 3.25LQEE216 pKa = 4.33AVSEE220 pKa = 4.55VEE222 pKa = 4.42TVADD226 pKa = 4.55DD227 pKa = 4.36LNEE230 pKa = 4.13ATSEE234 pKa = 3.92IDD236 pKa = 3.39TVATSITNVDD246 pKa = 3.46TVGNNIANVNAVAGNATNINAVNSNSSNINTVAGNNTNINTVAGANANITTLAGINANITTVAGINSNVSTVAGISSDD324 pKa = 3.44VTAVAGISSDD334 pKa = 3.49VQAVEE339 pKa = 4.18NIKK342 pKa = 11.3ANVTTVAGISSAVSNVSGISSAVSAVNSNSSNINAVNSNSANINTVASNNSNISTVAGVSSDD404 pKa = 3.32VTTVAGIASDD414 pKa = 3.68VSAVEE419 pKa = 4.64NIASNVTTVAGMSTAINTVNSNATNVNAVGGAIANVNNVGGSIANVNTVATNLASVNAFGEE480 pKa = 4.67TYY482 pKa = 10.14RR483 pKa = 11.84IASSAPTSSLNSGDD497 pKa = 4.82LYY499 pKa = 11.44FNTSTNVLNVYY510 pKa = 9.07GASGWQNAGSSVNGTSEE527 pKa = 3.99RR528 pKa = 11.84YY529 pKa = 9.77KK530 pKa = 10.63FVASGTPTTLTGNDD544 pKa = 3.41ANGNTLAYY552 pKa = 9.83DD553 pKa = 3.44AGFIDD558 pKa = 5.23VYY560 pKa = 11.43LNGIKK565 pKa = 9.16MVNGTDD571 pKa = 3.42VTVTSGSSIVFASALTNGDD590 pKa = 2.73IVEE593 pKa = 4.38AVTFGTFSVANMNASNLTSGTVPDD617 pKa = 3.78ARR619 pKa = 11.84ITGTYY624 pKa = 9.16TGITGLDD631 pKa = 3.22LTDD634 pKa = 3.31NSKK637 pKa = 10.17IRR639 pKa = 11.84LGTDD643 pKa = 2.78QMLEE647 pKa = 4.15LLEE650 pKa = 4.38HH651 pKa = 5.22TQEE654 pKa = 4.31LQDD657 pKa = 3.46
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.29TGNGSTTAYY17 pKa = 9.9SISYY21 pKa = 8.14TYY23 pKa = 10.76RR24 pKa = 11.84DD25 pKa = 3.59AADD28 pKa = 5.02LIVSVNGVATTAYY41 pKa = 9.98SLNAAGTTLTFDD53 pKa = 3.62SAPASSSAIEE63 pKa = 4.0IRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.24TSQTTRR73 pKa = 11.84LTDD76 pKa = 3.47YY77 pKa = 11.08AAGSVLTEE85 pKa = 4.12NDD87 pKa = 3.84LDD89 pKa = 4.08TDD91 pKa = 3.79STQAFFMGQEE101 pKa = 4.68AIDD104 pKa = 4.37DD105 pKa = 4.34ANDD108 pKa = 3.5VIKK111 pKa = 10.73ISSTDD116 pKa = 3.76FQFNAINKK124 pKa = 7.92QIRR127 pKa = 11.84NVANPTSNQDD137 pKa = 3.03VATKK141 pKa = 10.09HH142 pKa = 5.43YY143 pKa = 10.9LEE145 pKa = 4.34NTFLTTANKK154 pKa = 8.32TALTTINANIANIIAVNNNSTNINSAVSNATNINTVATNIGSVNTVAADD203 pKa = 3.22IAKK206 pKa = 10.13VIEE209 pKa = 4.24VANDD213 pKa = 3.25LQEE216 pKa = 4.33AVSEE220 pKa = 4.55VEE222 pKa = 4.42TVADD226 pKa = 4.55DD227 pKa = 4.36LNEE230 pKa = 4.13ATSEE234 pKa = 3.92IDD236 pKa = 3.39TVATSITNVDD246 pKa = 3.46TVGNNIANVNAVAGNATNINAVNSNSSNINTVAGNNTNINTVAGANANITTLAGINANITTVAGINSNVSTVAGISSDD324 pKa = 3.44VTAVAGISSDD334 pKa = 3.49VQAVEE339 pKa = 4.18NIKK342 pKa = 11.3ANVTTVAGISSAVSNVSGISSAVSAVNSNSSNINAVNSNSANINTVASNNSNISTVAGVSSDD404 pKa = 3.32VTTVAGIASDD414 pKa = 3.68VSAVEE419 pKa = 4.64NIASNVTTVAGMSTAINTVNSNATNVNAVGGAIANVNNVGGSIANVNTVATNLASVNAFGEE480 pKa = 4.67TYY482 pKa = 10.14RR483 pKa = 11.84IASSAPTSSLNSGDD497 pKa = 4.82LYY499 pKa = 11.44FNTSTNVLNVYY510 pKa = 9.07GASGWQNAGSSVNGTSEE527 pKa = 3.99RR528 pKa = 11.84YY529 pKa = 9.77KK530 pKa = 10.63FVASGTPTTLTGNDD544 pKa = 3.41ANGNTLAYY552 pKa = 9.83DD553 pKa = 3.44AGFIDD558 pKa = 5.23VYY560 pKa = 11.43LNGIKK565 pKa = 9.16MVNGTDD571 pKa = 3.42VTVTSGSSIVFASALTNGDD590 pKa = 2.73IVEE593 pKa = 4.38AVTFGTFSVANMNASNLTSGTVPDD617 pKa = 3.78ARR619 pKa = 11.84ITGTYY624 pKa = 9.16TGITGLDD631 pKa = 3.22LTDD634 pKa = 3.31NSKK637 pKa = 10.17IRR639 pKa = 11.84LGTDD643 pKa = 2.78QMLEE647 pKa = 4.15LLEE650 pKa = 4.38HH651 pKa = 5.22TQEE654 pKa = 4.31LQDD657 pKa = 3.46
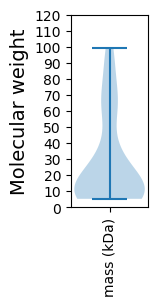
Molecular weight: 66.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NSY3|A0A4Y1NSY3_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC120P OX=2283021 GN=P120_gp35 PE=4 SV=1
MM1 pKa = 7.74AKK3 pKa = 10.23NGLYY7 pKa = 10.91ANIHH11 pKa = 6.08KK12 pKa = 10.25KK13 pKa = 9.21RR14 pKa = 11.84ARR16 pKa = 11.84IKK18 pKa = 10.26SGSGEE23 pKa = 4.08KK24 pKa = 9.51MRR26 pKa = 11.84TAGTKK31 pKa = 9.77GRR33 pKa = 11.84PTTAQFKK40 pKa = 10.05RR41 pKa = 11.84AAKK44 pKa = 8.06TAKK47 pKa = 10.08SS48 pKa = 3.41
MM1 pKa = 7.74AKK3 pKa = 10.23NGLYY7 pKa = 10.91ANIHH11 pKa = 6.08KK12 pKa = 10.25KK13 pKa = 9.21RR14 pKa = 11.84ARR16 pKa = 11.84IKK18 pKa = 10.26SGSGEE23 pKa = 4.08KK24 pKa = 9.51MRR26 pKa = 11.84TAGTKK31 pKa = 9.77GRR33 pKa = 11.84PTTAQFKK40 pKa = 10.05RR41 pKa = 11.84AAKK44 pKa = 8.06TAKK47 pKa = 10.08SS48 pKa = 3.41
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13103 |
44 |
875 |
247.2 |
27.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.983 ± 0.451 | 0.885 ± 0.169 |
5.899 ± 0.254 | 6.075 ± 0.396 |
4.098 ± 0.202 | 6.502 ± 0.384 |
1.702 ± 0.164 | 6.617 ± 0.209 |
8.113 ± 0.652 | 8.136 ± 0.387 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.25 | 6.724 ± 0.507 |
3.167 ± 0.218 | 3.969 ± 0.274 |
3.976 ± 0.329 | 7.243 ± 0.539 |
7.159 ± 0.554 | 5.983 ± 0.382 |
1.183 ± 0.127 | 3.16 ± 0.177 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
