
Hubei sobemo-like virus 24
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.48
Get precalculated fractions of proteins
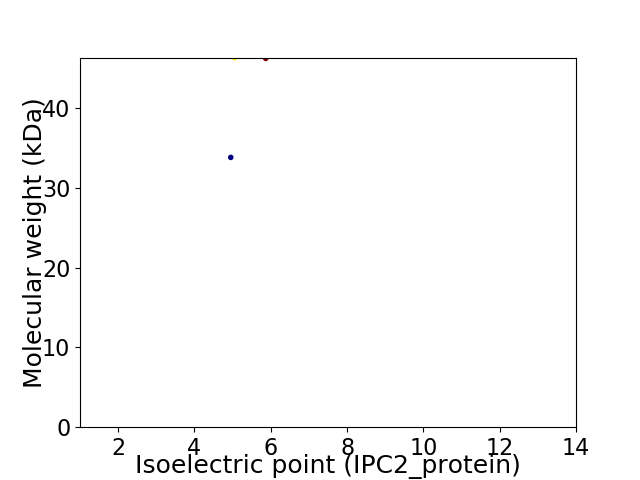
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEZ7|A0A1L3KEZ7_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 24 OX=1923210 PE=4 SV=1
MM1 pKa = 6.97TVEE4 pKa = 5.62DD5 pKa = 4.22FDD7 pKa = 4.55EE8 pKa = 4.88DD9 pKa = 5.14RR10 pKa = 11.84VDD12 pKa = 2.92SWAYY16 pKa = 10.36ADD18 pKa = 4.85QEE20 pKa = 3.91MDD22 pKa = 3.2YY23 pKa = 11.59DD24 pKa = 3.85RR25 pKa = 11.84VLDD28 pKa = 4.5FGDD31 pKa = 4.46DD32 pKa = 3.87SADD35 pKa = 3.25EE36 pKa = 4.55EE37 pKa = 4.77YY38 pKa = 10.99RR39 pKa = 11.84FDD41 pKa = 5.91DD42 pKa = 5.06RR43 pKa = 11.84DD44 pKa = 3.62DD45 pKa = 4.75DD46 pKa = 6.01RR47 pKa = 11.84DD48 pKa = 3.99DD49 pKa = 3.51RR50 pKa = 11.84WSDD53 pKa = 3.81DD54 pKa = 3.51EE55 pKa = 5.41LDD57 pKa = 3.91AYY59 pKa = 9.53EE60 pKa = 5.65DD61 pKa = 3.37KK62 pKa = 11.4YY63 pKa = 11.24EE64 pKa = 4.34RR65 pKa = 11.84NRR67 pKa = 11.84EE68 pKa = 3.78KK69 pKa = 10.49RR70 pKa = 11.84DD71 pKa = 3.4RR72 pKa = 11.84AEE74 pKa = 4.01EE75 pKa = 3.83RR76 pKa = 11.84WDD78 pKa = 3.65YY79 pKa = 11.82LNDD82 pKa = 3.88DD83 pKa = 3.26RR84 pKa = 11.84DD85 pKa = 3.96DD86 pKa = 3.95YY87 pKa = 11.8VRR89 pKa = 11.84EE90 pKa = 3.87ALGISRR96 pKa = 11.84MTPAQKK102 pKa = 10.44NRR104 pKa = 11.84LKK106 pKa = 10.95SLLDD110 pKa = 3.42RR111 pKa = 11.84PLVVKK116 pKa = 10.06GQAPGAPNVEE126 pKa = 4.38HH127 pKa = 7.07PPPTEE132 pKa = 3.8EE133 pKa = 4.69LDD135 pKa = 3.52LDD137 pKa = 3.98EE138 pKa = 5.0VVGNLVEE145 pKa = 4.37RR146 pKa = 11.84VEE148 pKa = 4.11HH149 pKa = 7.02LEE151 pKa = 4.61AEE153 pKa = 4.26LANLKK158 pKa = 10.56GKK160 pKa = 10.24LIPKK164 pKa = 9.88KK165 pKa = 10.23GAPPVKK171 pKa = 10.05KK172 pKa = 9.78PKK174 pKa = 10.22KK175 pKa = 9.33CVHH178 pKa = 6.15CSEE181 pKa = 4.29MLEE184 pKa = 4.43SIAAAAKK191 pKa = 9.77HH192 pKa = 5.82RR193 pKa = 11.84RR194 pKa = 11.84EE195 pKa = 4.13QHH197 pKa = 6.18PEE199 pKa = 3.8AYY201 pKa = 8.36WAARR205 pKa = 11.84NATEE209 pKa = 3.81NPGYY213 pKa = 10.24KK214 pKa = 10.29GEE216 pKa = 4.33SANGADD222 pKa = 3.4FKK224 pKa = 11.71VRR226 pKa = 11.84VKK228 pKa = 9.58TQAKK232 pKa = 8.58PAFFRR237 pKa = 11.84GGSRR241 pKa = 11.84KK242 pKa = 7.28TSKK245 pKa = 10.09PSSVSSSTTSGSSSGSPSPQEE266 pKa = 3.87TRR268 pKa = 11.84KK269 pKa = 8.52STNGQNQSDD278 pKa = 3.69DD279 pKa = 4.04RR280 pKa = 11.84FDD282 pKa = 4.29KK283 pKa = 10.7FLQAIIGLASEE294 pKa = 5.05MKK296 pKa = 10.45QKK298 pKa = 11.01
MM1 pKa = 6.97TVEE4 pKa = 5.62DD5 pKa = 4.22FDD7 pKa = 4.55EE8 pKa = 4.88DD9 pKa = 5.14RR10 pKa = 11.84VDD12 pKa = 2.92SWAYY16 pKa = 10.36ADD18 pKa = 4.85QEE20 pKa = 3.91MDD22 pKa = 3.2YY23 pKa = 11.59DD24 pKa = 3.85RR25 pKa = 11.84VLDD28 pKa = 4.5FGDD31 pKa = 4.46DD32 pKa = 3.87SADD35 pKa = 3.25EE36 pKa = 4.55EE37 pKa = 4.77YY38 pKa = 10.99RR39 pKa = 11.84FDD41 pKa = 5.91DD42 pKa = 5.06RR43 pKa = 11.84DD44 pKa = 3.62DD45 pKa = 4.75DD46 pKa = 6.01RR47 pKa = 11.84DD48 pKa = 3.99DD49 pKa = 3.51RR50 pKa = 11.84WSDD53 pKa = 3.81DD54 pKa = 3.51EE55 pKa = 5.41LDD57 pKa = 3.91AYY59 pKa = 9.53EE60 pKa = 5.65DD61 pKa = 3.37KK62 pKa = 11.4YY63 pKa = 11.24EE64 pKa = 4.34RR65 pKa = 11.84NRR67 pKa = 11.84EE68 pKa = 3.78KK69 pKa = 10.49RR70 pKa = 11.84DD71 pKa = 3.4RR72 pKa = 11.84AEE74 pKa = 4.01EE75 pKa = 3.83RR76 pKa = 11.84WDD78 pKa = 3.65YY79 pKa = 11.82LNDD82 pKa = 3.88DD83 pKa = 3.26RR84 pKa = 11.84DD85 pKa = 3.96DD86 pKa = 3.95YY87 pKa = 11.8VRR89 pKa = 11.84EE90 pKa = 3.87ALGISRR96 pKa = 11.84MTPAQKK102 pKa = 10.44NRR104 pKa = 11.84LKK106 pKa = 10.95SLLDD110 pKa = 3.42RR111 pKa = 11.84PLVVKK116 pKa = 10.06GQAPGAPNVEE126 pKa = 4.38HH127 pKa = 7.07PPPTEE132 pKa = 3.8EE133 pKa = 4.69LDD135 pKa = 3.52LDD137 pKa = 3.98EE138 pKa = 5.0VVGNLVEE145 pKa = 4.37RR146 pKa = 11.84VEE148 pKa = 4.11HH149 pKa = 7.02LEE151 pKa = 4.61AEE153 pKa = 4.26LANLKK158 pKa = 10.56GKK160 pKa = 10.24LIPKK164 pKa = 9.88KK165 pKa = 10.23GAPPVKK171 pKa = 10.05KK172 pKa = 9.78PKK174 pKa = 10.22KK175 pKa = 9.33CVHH178 pKa = 6.15CSEE181 pKa = 4.29MLEE184 pKa = 4.43SIAAAAKK191 pKa = 9.77HH192 pKa = 5.82RR193 pKa = 11.84RR194 pKa = 11.84EE195 pKa = 4.13QHH197 pKa = 6.18PEE199 pKa = 3.8AYY201 pKa = 8.36WAARR205 pKa = 11.84NATEE209 pKa = 3.81NPGYY213 pKa = 10.24KK214 pKa = 10.29GEE216 pKa = 4.33SANGADD222 pKa = 3.4FKK224 pKa = 11.71VRR226 pKa = 11.84VKK228 pKa = 9.58TQAKK232 pKa = 8.58PAFFRR237 pKa = 11.84GGSRR241 pKa = 11.84KK242 pKa = 7.28TSKK245 pKa = 10.09PSSVSSSTTSGSSSGSPSPQEE266 pKa = 3.87TRR268 pKa = 11.84KK269 pKa = 8.52STNGQNQSDD278 pKa = 3.69DD279 pKa = 4.04RR280 pKa = 11.84FDD282 pKa = 4.29KK283 pKa = 10.7FLQAIIGLASEE294 pKa = 5.05MKK296 pKa = 10.45QKK298 pKa = 11.01
Molecular weight: 33.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEZ7|A0A1L3KEZ7_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 24 OX=1923210 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.39SLAIHH7 pKa = 6.01SAMHH11 pKa = 6.2KK12 pKa = 10.24GLRR15 pKa = 11.84TDD17 pKa = 3.73PPSDD21 pKa = 3.63VQEE24 pKa = 4.55DD25 pKa = 4.68EE26 pKa = 3.91ILEE29 pKa = 4.17WMARR33 pKa = 11.84IYY35 pKa = 9.94PRR37 pKa = 11.84WEE39 pKa = 3.8IPSDD43 pKa = 3.48FMQYY47 pKa = 7.3THH49 pKa = 5.89YY50 pKa = 10.81QRR52 pKa = 11.84VLRR55 pKa = 11.84GLDD58 pKa = 3.24WTSSPGYY65 pKa = 9.26PYY67 pKa = 11.51LLTSPTNSQLFQVVDD82 pKa = 4.11GEE84 pKa = 4.26PAEE87 pKa = 4.38APSRR91 pKa = 11.84AIWEE95 pKa = 4.28IVQKK99 pKa = 10.88RR100 pKa = 11.84LVEE103 pKa = 4.13RR104 pKa = 11.84DD105 pKa = 3.21ADD107 pKa = 4.39PIRR110 pKa = 11.84LFVKK114 pKa = 10.21PEE116 pKa = 3.47PHH118 pKa = 6.48KK119 pKa = 10.75LKK121 pKa = 10.53KK122 pKa = 9.84IEE124 pKa = 3.93EE125 pKa = 4.2GRR127 pKa = 11.84FRR129 pKa = 11.84LISSVSVVDD138 pKa = 4.52QIIDD142 pKa = 3.47HH143 pKa = 6.38MLHH146 pKa = 7.32DD147 pKa = 6.13DD148 pKa = 3.73YY149 pKa = 11.63DD150 pKa = 3.65QVMIEE155 pKa = 3.98NWATVPPKK163 pKa = 10.79AGWTPLLGGWKK174 pKa = 8.18TVPNFLRR181 pKa = 11.84PLAVDD186 pKa = 3.83KK187 pKa = 11.07KK188 pKa = 9.42AWDD191 pKa = 3.28WTVRR195 pKa = 11.84PWLLSLSLRR204 pKa = 11.84HH205 pKa = 7.14RR206 pKa = 11.84IRR208 pKa = 11.84QCSNMCPLWEE218 pKa = 5.65DD219 pKa = 3.08LAEE222 pKa = 4.16WRR224 pKa = 11.84WRR226 pKa = 11.84MLYY229 pKa = 10.69AEE231 pKa = 4.63AVFVTSGGILLKK243 pKa = 10.55QKK245 pKa = 10.44EE246 pKa = 4.15PGVRR250 pKa = 11.84KK251 pKa = 9.85SGCVTTLSDD260 pKa = 3.98NSLEE264 pKa = 3.76QDD266 pKa = 2.94ILHH269 pKa = 6.37RR270 pKa = 11.84RR271 pKa = 11.84ISLDD275 pKa = 2.98MWRR278 pKa = 11.84KK279 pKa = 9.62NPDD282 pKa = 3.05LHH284 pKa = 7.55IPPLWGSMGDD294 pKa = 3.52DD295 pKa = 4.52TIQEE299 pKa = 4.23EE300 pKa = 4.52PDD302 pKa = 3.45NLDD305 pKa = 3.14EE306 pKa = 4.17YY307 pKa = 11.3LGYY310 pKa = 10.7LSQYY314 pKa = 9.86CIVKK318 pKa = 9.92EE319 pKa = 4.16AVRR322 pKa = 11.84SPDD325 pKa = 3.14FAGFSFHH332 pKa = 6.92GRR334 pKa = 11.84DD335 pKa = 3.6VKK337 pKa = 10.62PLYY340 pKa = 9.96LGKK343 pKa = 9.65HH344 pKa = 6.22AYY346 pKa = 9.23QLLHH350 pKa = 6.64VDD352 pKa = 3.79PAVKK356 pKa = 10.5DD357 pKa = 3.99GVARR361 pKa = 11.84SYY363 pKa = 11.74ALLYY367 pKa = 10.37HH368 pKa = 7.11RR369 pKa = 11.84APAARR374 pKa = 11.84WISRR378 pKa = 11.84MCRR381 pKa = 11.84MLGDD385 pKa = 3.42VPTEE389 pKa = 3.78VRR391 pKa = 11.84LDD393 pKa = 3.7SIYY396 pKa = 11.08DD397 pKa = 3.6GEE399 pKa = 4.23
MM1 pKa = 7.6KK2 pKa = 10.39SLAIHH7 pKa = 6.01SAMHH11 pKa = 6.2KK12 pKa = 10.24GLRR15 pKa = 11.84TDD17 pKa = 3.73PPSDD21 pKa = 3.63VQEE24 pKa = 4.55DD25 pKa = 4.68EE26 pKa = 3.91ILEE29 pKa = 4.17WMARR33 pKa = 11.84IYY35 pKa = 9.94PRR37 pKa = 11.84WEE39 pKa = 3.8IPSDD43 pKa = 3.48FMQYY47 pKa = 7.3THH49 pKa = 5.89YY50 pKa = 10.81QRR52 pKa = 11.84VLRR55 pKa = 11.84GLDD58 pKa = 3.24WTSSPGYY65 pKa = 9.26PYY67 pKa = 11.51LLTSPTNSQLFQVVDD82 pKa = 4.11GEE84 pKa = 4.26PAEE87 pKa = 4.38APSRR91 pKa = 11.84AIWEE95 pKa = 4.28IVQKK99 pKa = 10.88RR100 pKa = 11.84LVEE103 pKa = 4.13RR104 pKa = 11.84DD105 pKa = 3.21ADD107 pKa = 4.39PIRR110 pKa = 11.84LFVKK114 pKa = 10.21PEE116 pKa = 3.47PHH118 pKa = 6.48KK119 pKa = 10.75LKK121 pKa = 10.53KK122 pKa = 9.84IEE124 pKa = 3.93EE125 pKa = 4.2GRR127 pKa = 11.84FRR129 pKa = 11.84LISSVSVVDD138 pKa = 4.52QIIDD142 pKa = 3.47HH143 pKa = 6.38MLHH146 pKa = 7.32DD147 pKa = 6.13DD148 pKa = 3.73YY149 pKa = 11.63DD150 pKa = 3.65QVMIEE155 pKa = 3.98NWATVPPKK163 pKa = 10.79AGWTPLLGGWKK174 pKa = 8.18TVPNFLRR181 pKa = 11.84PLAVDD186 pKa = 3.83KK187 pKa = 11.07KK188 pKa = 9.42AWDD191 pKa = 3.28WTVRR195 pKa = 11.84PWLLSLSLRR204 pKa = 11.84HH205 pKa = 7.14RR206 pKa = 11.84IRR208 pKa = 11.84QCSNMCPLWEE218 pKa = 5.65DD219 pKa = 3.08LAEE222 pKa = 4.16WRR224 pKa = 11.84WRR226 pKa = 11.84MLYY229 pKa = 10.69AEE231 pKa = 4.63AVFVTSGGILLKK243 pKa = 10.55QKK245 pKa = 10.44EE246 pKa = 4.15PGVRR250 pKa = 11.84KK251 pKa = 9.85SGCVTTLSDD260 pKa = 3.98NSLEE264 pKa = 3.76QDD266 pKa = 2.94ILHH269 pKa = 6.37RR270 pKa = 11.84RR271 pKa = 11.84ISLDD275 pKa = 2.98MWRR278 pKa = 11.84KK279 pKa = 9.62NPDD282 pKa = 3.05LHH284 pKa = 7.55IPPLWGSMGDD294 pKa = 3.52DD295 pKa = 4.52TIQEE299 pKa = 4.23EE300 pKa = 4.52PDD302 pKa = 3.45NLDD305 pKa = 3.14EE306 pKa = 4.17YY307 pKa = 11.3LGYY310 pKa = 10.7LSQYY314 pKa = 9.86CIVKK318 pKa = 9.92EE319 pKa = 4.16AVRR322 pKa = 11.84SPDD325 pKa = 3.14FAGFSFHH332 pKa = 6.92GRR334 pKa = 11.84DD335 pKa = 3.6VKK337 pKa = 10.62PLYY340 pKa = 9.96LGKK343 pKa = 9.65HH344 pKa = 6.22AYY346 pKa = 9.23QLLHH350 pKa = 6.64VDD352 pKa = 3.79PAVKK356 pKa = 10.5DD357 pKa = 3.99GVARR361 pKa = 11.84SYY363 pKa = 11.74ALLYY367 pKa = 10.37HH368 pKa = 7.11RR369 pKa = 11.84APAARR374 pKa = 11.84WISRR378 pKa = 11.84MCRR381 pKa = 11.84MLGDD385 pKa = 3.42VPTEE389 pKa = 3.78VRR391 pKa = 11.84LDD393 pKa = 3.7SIYY396 pKa = 11.08DD397 pKa = 3.6GEE399 pKa = 4.23
Molecular weight: 46.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
697 |
298 |
399 |
348.5 |
40.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.03 ± 1.075 | 1.004 ± 0.211 |
9.469 ± 1.444 | 7.317 ± 1.319 |
2.439 ± 0.156 | 5.308 ± 0.038 |
2.582 ± 0.574 | 3.73 ± 1.302 |
6.169 ± 1.196 | 8.752 ± 1.72 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.439 ± 0.483 | 2.582 ± 0.703 |
6.6 ± 0.568 | 3.443 ± 0.056 |
7.891 ± 0.316 | 7.317 ± 0.467 |
3.443 ± 0.056 | 6.169 ± 0.508 |
2.869 ± 0.969 | 3.443 ± 0.268 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
