
Lentibacter virus vB_LenP_ICBM3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Zobellviridae; Cobavirinae; Siovirus; unclassified Siovirus
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
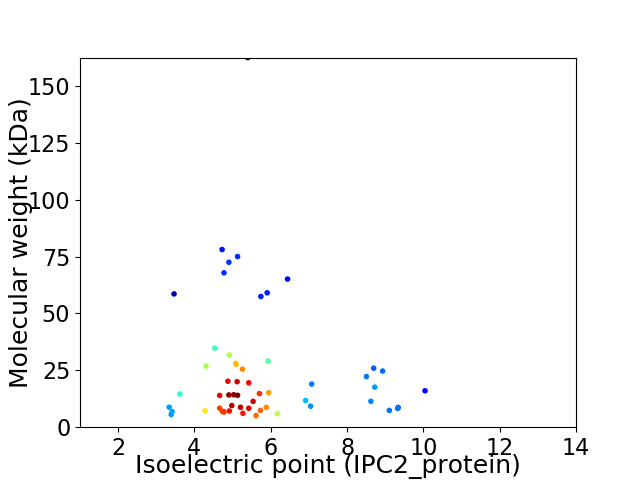
Virtual 2D-PAGE plot for 56 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G2YRD4|A0A3G2YRD4_9CAUD Uncharacterized protein OS=Lentibacter virus vB_LenP_ICBM3 OX=2301530 PE=4 SV=1
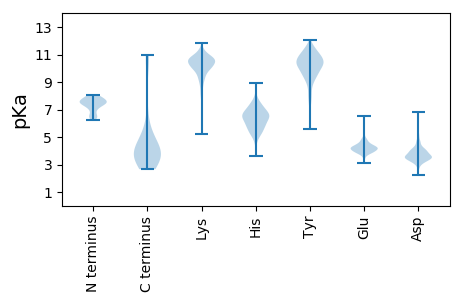
MM1 pKa = 7.61AKK3 pKa = 10.08KK4 pKa = 8.7PTISTLQSGFNSTEE18 pKa = 3.89TLNSNFEE25 pKa = 4.21KK26 pKa = 10.87LRR28 pKa = 11.84DD29 pKa = 3.81ALDD32 pKa = 3.53NTLSLDD38 pKa = 3.43GSTPNAMEE46 pKa = 5.54ADD48 pKa = 3.65LDD50 pKa = 4.43LNGNNIIGATGLLINGTDD68 pKa = 3.58YY69 pKa = 11.52LIDD72 pKa = 3.63VEE74 pKa = 4.32AAKK77 pKa = 10.41AAALVAQAAAEE88 pKa = 3.93LAEE91 pKa = 4.56NNAEE95 pKa = 4.13TAEE98 pKa = 4.25VNAEE102 pKa = 3.92ASEE105 pKa = 4.25TAAGLLATAASTSATNAATSATAASVSEE133 pKa = 4.08TAAALSEE140 pKa = 4.43TNAATSEE147 pKa = 4.57TNASTSEE154 pKa = 4.06TNAAASALAASTSEE168 pKa = 4.09TNAASSEE175 pKa = 4.29TNAAASEE182 pKa = 4.43TAASTSEE189 pKa = 4.04TNAAASEE196 pKa = 4.43TAASTSEE203 pKa = 4.13TNAATSEE210 pKa = 4.33TNAATSEE217 pKa = 4.26TNAAASEE224 pKa = 4.21AASSASAAAALTSEE238 pKa = 4.28TAAALSEE245 pKa = 4.38ANASASEE252 pKa = 3.94AAAGLSEE259 pKa = 4.04TAAAASEE266 pKa = 4.09TAAASSASSASTSASNAASSASSAQASKK294 pKa = 11.0DD295 pKa = 3.4AALAALDD302 pKa = 4.23SFDD305 pKa = 5.84DD306 pKa = 4.2RR307 pKa = 11.84YY308 pKa = 11.02LGQKK312 pKa = 10.02VSDD315 pKa = 3.76PTLDD319 pKa = 3.44NDD321 pKa = 4.31GDD323 pKa = 4.21ALVAGALYY331 pKa = 10.66FNTTDD336 pKa = 5.31DD337 pKa = 3.03IMKK340 pKa = 9.94VYY342 pKa = 10.29EE343 pKa = 4.1GSVWVAAYY351 pKa = 10.26ASLSGALLANNNLSDD366 pKa = 4.16LANVSATRR374 pKa = 11.84TNLGLGTAATTASTDD389 pKa = 3.38YY390 pKa = 9.88ATAAQGSLADD400 pKa = 4.33SAIQPADD407 pKa = 3.51LATVATTGVYY417 pKa = 10.7ADD419 pKa = 4.07LTGKK423 pKa = 7.69PTLGTAAATDD433 pKa = 3.58STAYY437 pKa = 9.06ATAAQGALADD447 pKa = 4.12SAIQAADD454 pKa = 3.64LATVATTGAYY464 pKa = 10.05SDD466 pKa = 4.02LTGTPAPYY474 pKa = 10.84ADD476 pKa = 4.31ADD478 pKa = 3.77VDD480 pKa = 3.77THH482 pKa = 6.98LNTGTAATGEE492 pKa = 4.19YY493 pKa = 10.36LSWNGADD500 pKa = 3.98YY501 pKa = 11.23DD502 pKa = 4.07WATVPAGYY510 pKa = 10.87ADD512 pKa = 4.3ADD514 pKa = 3.69VDD516 pKa = 3.79THH518 pKa = 6.99LNTGTATTDD527 pKa = 3.59QVLAWTGSDD536 pKa = 3.88YY537 pKa = 11.79DD538 pKa = 3.85WVDD541 pKa = 3.43ASAGATGGGTDD552 pKa = 4.65KK553 pKa = 11.05IFVEE557 pKa = 4.27NGQVVTTNYY566 pKa = 9.03TVAATTNAMTTGPIDD581 pKa = 3.43INSGVTVTVEE591 pKa = 3.66TGGRR595 pKa = 11.84WVVII599 pKa = 4.05
MM1 pKa = 7.61AKK3 pKa = 10.08KK4 pKa = 8.7PTISTLQSGFNSTEE18 pKa = 3.89TLNSNFEE25 pKa = 4.21KK26 pKa = 10.87LRR28 pKa = 11.84DD29 pKa = 3.81ALDD32 pKa = 3.53NTLSLDD38 pKa = 3.43GSTPNAMEE46 pKa = 5.54ADD48 pKa = 3.65LDD50 pKa = 4.43LNGNNIIGATGLLINGTDD68 pKa = 3.58YY69 pKa = 11.52LIDD72 pKa = 3.63VEE74 pKa = 4.32AAKK77 pKa = 10.41AAALVAQAAAEE88 pKa = 3.93LAEE91 pKa = 4.56NNAEE95 pKa = 4.13TAEE98 pKa = 4.25VNAEE102 pKa = 3.92ASEE105 pKa = 4.25TAAGLLATAASTSATNAATSATAASVSEE133 pKa = 4.08TAAALSEE140 pKa = 4.43TNAATSEE147 pKa = 4.57TNASTSEE154 pKa = 4.06TNAAASALAASTSEE168 pKa = 4.09TNAASSEE175 pKa = 4.29TNAAASEE182 pKa = 4.43TAASTSEE189 pKa = 4.04TNAAASEE196 pKa = 4.43TAASTSEE203 pKa = 4.13TNAATSEE210 pKa = 4.33TNAATSEE217 pKa = 4.26TNAAASEE224 pKa = 4.21AASSASAAAALTSEE238 pKa = 4.28TAAALSEE245 pKa = 4.38ANASASEE252 pKa = 3.94AAAGLSEE259 pKa = 4.04TAAAASEE266 pKa = 4.09TAAASSASSASTSASNAASSASSAQASKK294 pKa = 11.0DD295 pKa = 3.4AALAALDD302 pKa = 4.23SFDD305 pKa = 5.84DD306 pKa = 4.2RR307 pKa = 11.84YY308 pKa = 11.02LGQKK312 pKa = 10.02VSDD315 pKa = 3.76PTLDD319 pKa = 3.44NDD321 pKa = 4.31GDD323 pKa = 4.21ALVAGALYY331 pKa = 10.66FNTTDD336 pKa = 5.31DD337 pKa = 3.03IMKK340 pKa = 9.94VYY342 pKa = 10.29EE343 pKa = 4.1GSVWVAAYY351 pKa = 10.26ASLSGALLANNNLSDD366 pKa = 4.16LANVSATRR374 pKa = 11.84TNLGLGTAATTASTDD389 pKa = 3.38YY390 pKa = 9.88ATAAQGSLADD400 pKa = 4.33SAIQPADD407 pKa = 3.51LATVATTGVYY417 pKa = 10.7ADD419 pKa = 4.07LTGKK423 pKa = 7.69PTLGTAAATDD433 pKa = 3.58STAYY437 pKa = 9.06ATAAQGALADD447 pKa = 4.12SAIQAADD454 pKa = 3.64LATVATTGAYY464 pKa = 10.05SDD466 pKa = 4.02LTGTPAPYY474 pKa = 10.84ADD476 pKa = 4.31ADD478 pKa = 3.77VDD480 pKa = 3.77THH482 pKa = 6.98LNTGTAATGEE492 pKa = 4.19YY493 pKa = 10.36LSWNGADD500 pKa = 3.98YY501 pKa = 11.23DD502 pKa = 4.07WATVPAGYY510 pKa = 10.87ADD512 pKa = 4.3ADD514 pKa = 3.69VDD516 pKa = 3.79THH518 pKa = 6.99LNTGTATTDD527 pKa = 3.59QVLAWTGSDD536 pKa = 3.88YY537 pKa = 11.79DD538 pKa = 3.85WVDD541 pKa = 3.43ASAGATGGGTDD552 pKa = 4.65KK553 pKa = 11.05IFVEE557 pKa = 4.27NGQVVTTNYY566 pKa = 9.03TVAATTNAMTTGPIDD581 pKa = 3.43INSGVTVTVEE591 pKa = 3.66TGGRR595 pKa = 11.84WVVII599 pKa = 4.05
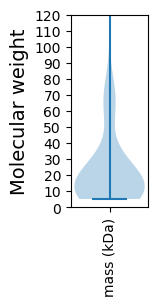
Molecular weight: 58.6 kDa
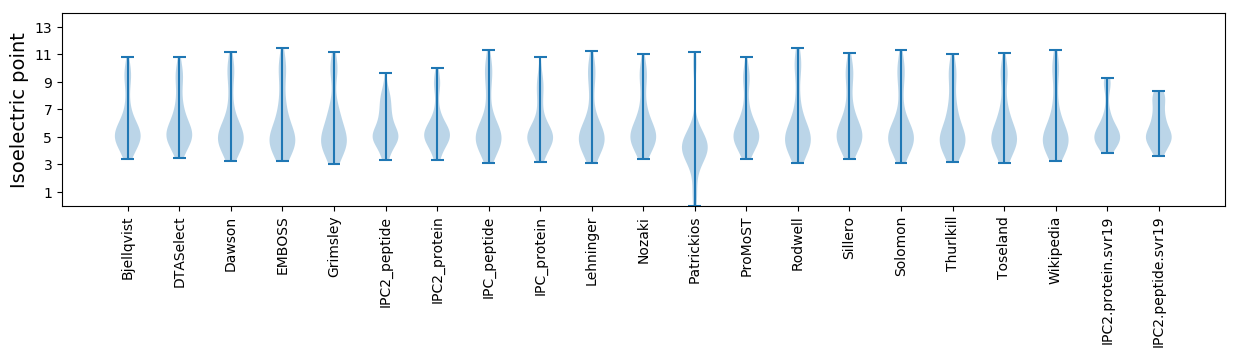
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2YRN2|A0A3G2YRN2_9CAUD Uncharacterized protein OS=Lentibacter virus vB_LenP_ICBM3 OX=2301530 PE=4 SV=1
MM1 pKa = 7.87AIFTAIGTAVLGAAAATAGAAATAIVGGVVAAGAVAGIAGTVKK44 pKa = 10.41AGKK47 pKa = 8.56ATRR50 pKa = 11.84KK51 pKa = 9.44AAAAQQKK58 pKa = 9.65AQEE61 pKa = 4.31LQSKK65 pKa = 7.44RR66 pKa = 11.84QRR68 pKa = 11.84RR69 pKa = 11.84AAIRR73 pKa = 11.84SNILASARR81 pKa = 11.84ARR83 pKa = 11.84ASAQAAGTAQSSGLSGAIGAGRR105 pKa = 11.84SQLGAEE111 pKa = 4.56LGFGTQMSGLSANISKK127 pKa = 10.64FNMQAQTYY135 pKa = 10.35GDD137 pKa = 3.43IAKK140 pKa = 10.39LGFAAAGNPTAIADD154 pKa = 5.08PIAGVYY160 pKa = 10.19DD161 pKa = 3.96YY162 pKa = 11.44LKK164 pKa = 10.75KK165 pKa = 10.9
MM1 pKa = 7.87AIFTAIGTAVLGAAAATAGAAATAIVGGVVAAGAVAGIAGTVKK44 pKa = 10.41AGKK47 pKa = 8.56ATRR50 pKa = 11.84KK51 pKa = 9.44AAAAQQKK58 pKa = 9.65AQEE61 pKa = 4.31LQSKK65 pKa = 7.44RR66 pKa = 11.84QRR68 pKa = 11.84RR69 pKa = 11.84AAIRR73 pKa = 11.84SNILASARR81 pKa = 11.84ARR83 pKa = 11.84ASAQAAGTAQSSGLSGAIGAGRR105 pKa = 11.84SQLGAEE111 pKa = 4.56LGFGTQMSGLSANISKK127 pKa = 10.64FNMQAQTYY135 pKa = 10.35GDD137 pKa = 3.43IAKK140 pKa = 10.39LGFAAAGNPTAIADD154 pKa = 5.08PIAGVYY160 pKa = 10.19DD161 pKa = 3.96YY162 pKa = 11.44LKK164 pKa = 10.75KK165 pKa = 10.9
Molecular weight: 16.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12523 |
46 |
1487 |
223.6 |
24.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.902 ± 0.952 | 0.711 ± 0.161 |
6.484 ± 0.232 | 6.843 ± 0.313 |
3.673 ± 0.269 | 7.858 ± 0.738 |
1.493 ± 0.159 | 4.839 ± 0.28 |
5.989 ± 0.47 | 8.049 ± 0.259 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.611 ± 0.187 | 4.576 ± 0.274 |
3.426 ± 0.205 | 3.569 ± 0.21 |
4.584 ± 0.306 | 6.851 ± 0.373 |
6.923 ± 0.483 | 6.636 ± 0.277 |
1.437 ± 0.167 | 3.545 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
