
Echinococcus granulosus (Hydatid tapeworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Cestoda; Eucestoda; Cyclophyllidea; Taeniidae; Echinococcus;
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
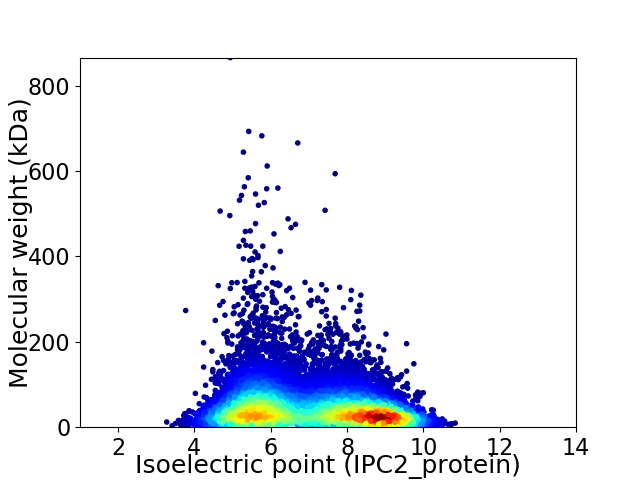
Virtual 2D-PAGE plot for 11279 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W6U0T9|W6U0T9_ECHGR Uncharacterized protein OS=Echinococcus granulosus OX=6210 GN=EGR_10476 PE=4 SV=1
MM1 pKa = 7.08KK2 pKa = 9.75TLLFLLTLSAILLVACTKK20 pKa = 10.26ARR22 pKa = 11.84PYY24 pKa = 6.71TTPIATQLRR33 pKa = 11.84DD34 pKa = 3.51EE35 pKa = 4.79RR36 pKa = 11.84GEE38 pKa = 3.96EE39 pKa = 4.2SSGKK43 pKa = 10.17DD44 pKa = 3.07AVSQADD50 pKa = 3.52GHH52 pKa = 7.05DD53 pKa = 3.65EE54 pKa = 3.9QDD56 pKa = 3.51DD57 pKa = 4.05NEE59 pKa = 5.3DD60 pKa = 3.64YY61 pKa = 11.42DD62 pKa = 4.08VFIYY66 pKa = 10.55DD67 pKa = 4.37SYY69 pKa = 11.76NDD71 pKa = 3.83TVKK74 pKa = 11.08ADD76 pKa = 3.53AQEE79 pKa = 4.2VEE81 pKa = 4.28EE82 pKa = 4.47TEE84 pKa = 3.99AAEE87 pKa = 4.71EE88 pKa = 4.01YY89 pKa = 11.02DD90 pKa = 3.19EE91 pKa = 6.3AYY93 pKa = 11.08YY94 pKa = 11.0DD95 pKa = 3.96DD96 pKa = 4.04EE97 pKa = 4.74WYY99 pKa = 10.78AKK101 pKa = 9.28YY102 pKa = 10.86APVYY106 pKa = 11.02VDD108 pKa = 4.69FEE110 pKa = 4.54EE111 pKa = 5.06KK112 pKa = 10.66KK113 pKa = 10.96EE114 pKa = 3.95GDD116 pKa = 3.32KK117 pKa = 11.36DD118 pKa = 4.18KK119 pKa = 11.24EE120 pKa = 4.17DD121 pKa = 3.64EE122 pKa = 4.41RR123 pKa = 11.84EE124 pKa = 4.0GLTFDD129 pKa = 3.78DD130 pKa = 5.07ARR132 pKa = 11.84YY133 pKa = 6.64TQHH136 pKa = 5.58TPAYY140 pKa = 8.83KK141 pKa = 10.27EE142 pKa = 3.97EE143 pKa = 4.2EE144 pKa = 4.25EE145 pKa = 4.66KK146 pKa = 11.3EE147 pKa = 3.98EE148 pKa = 4.07TEE150 pKa = 3.95AAEE153 pKa = 4.78EE154 pKa = 4.01YY155 pKa = 11.02DD156 pKa = 3.19EE157 pKa = 6.3AYY159 pKa = 11.08YY160 pKa = 11.0DD161 pKa = 3.96DD162 pKa = 4.04EE163 pKa = 4.74WYY165 pKa = 10.78AKK167 pKa = 9.15YY168 pKa = 10.81APVYY172 pKa = 9.12VQSDD176 pKa = 3.93EE177 pKa = 4.52NEE179 pKa = 4.07EE180 pKa = 3.99
MM1 pKa = 7.08KK2 pKa = 9.75TLLFLLTLSAILLVACTKK20 pKa = 10.26ARR22 pKa = 11.84PYY24 pKa = 6.71TTPIATQLRR33 pKa = 11.84DD34 pKa = 3.51EE35 pKa = 4.79RR36 pKa = 11.84GEE38 pKa = 3.96EE39 pKa = 4.2SSGKK43 pKa = 10.17DD44 pKa = 3.07AVSQADD50 pKa = 3.52GHH52 pKa = 7.05DD53 pKa = 3.65EE54 pKa = 3.9QDD56 pKa = 3.51DD57 pKa = 4.05NEE59 pKa = 5.3DD60 pKa = 3.64YY61 pKa = 11.42DD62 pKa = 4.08VFIYY66 pKa = 10.55DD67 pKa = 4.37SYY69 pKa = 11.76NDD71 pKa = 3.83TVKK74 pKa = 11.08ADD76 pKa = 3.53AQEE79 pKa = 4.2VEE81 pKa = 4.28EE82 pKa = 4.47TEE84 pKa = 3.99AAEE87 pKa = 4.71EE88 pKa = 4.01YY89 pKa = 11.02DD90 pKa = 3.19EE91 pKa = 6.3AYY93 pKa = 11.08YY94 pKa = 11.0DD95 pKa = 3.96DD96 pKa = 4.04EE97 pKa = 4.74WYY99 pKa = 10.78AKK101 pKa = 9.28YY102 pKa = 10.86APVYY106 pKa = 11.02VDD108 pKa = 4.69FEE110 pKa = 4.54EE111 pKa = 5.06KK112 pKa = 10.66KK113 pKa = 10.96EE114 pKa = 3.95GDD116 pKa = 3.32KK117 pKa = 11.36DD118 pKa = 4.18KK119 pKa = 11.24EE120 pKa = 4.17DD121 pKa = 3.64EE122 pKa = 4.41RR123 pKa = 11.84EE124 pKa = 4.0GLTFDD129 pKa = 3.78DD130 pKa = 5.07ARR132 pKa = 11.84YY133 pKa = 6.64TQHH136 pKa = 5.58TPAYY140 pKa = 8.83KK141 pKa = 10.27EE142 pKa = 3.97EE143 pKa = 4.2EE144 pKa = 4.25EE145 pKa = 4.66KK146 pKa = 11.3EE147 pKa = 3.98EE148 pKa = 4.07TEE150 pKa = 3.95AAEE153 pKa = 4.78EE154 pKa = 4.01YY155 pKa = 11.02DD156 pKa = 3.19EE157 pKa = 6.3AYY159 pKa = 11.08YY160 pKa = 11.0DD161 pKa = 3.96DD162 pKa = 4.04EE163 pKa = 4.74WYY165 pKa = 10.78AKK167 pKa = 9.15YY168 pKa = 10.81APVYY172 pKa = 9.12VQSDD176 pKa = 3.93EE177 pKa = 4.52NEE179 pKa = 4.07EE180 pKa = 3.99
Molecular weight: 21.06 kDa
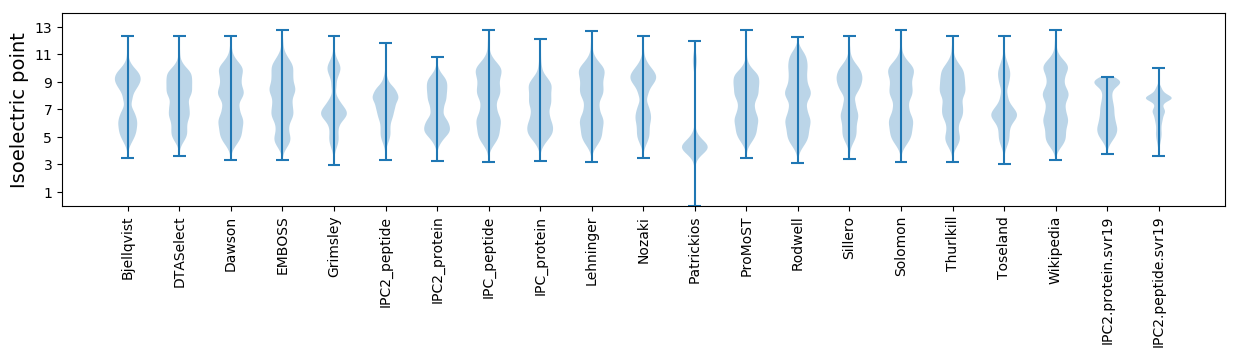
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6UE18|W6UE18_ECHGR Uncharacterized protein OS=Echinococcus granulosus OX=6210 GN=EGR_05894 PE=4 SV=1
MM1 pKa = 7.45ALHH4 pKa = 6.78RR5 pKa = 11.84RR6 pKa = 11.84LHH8 pKa = 6.06PQPQPPMRR16 pKa = 11.84SPPFRR21 pKa = 11.84PSRR24 pKa = 11.84LPPSTFPSALDD35 pKa = 3.56LTLSLALHH43 pKa = 6.53TCTHH47 pKa = 6.76AGRR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84SRR55 pKa = 11.84SHH57 pKa = 4.92NTPRR61 pKa = 11.84LAPFSQHH68 pKa = 5.7VLLLASRR75 pKa = 11.84SLSCAYY81 pKa = 9.77CQQAVATSSQWLSARR96 pKa = 11.84VSAVRR101 pKa = 11.84SCALGRR107 pKa = 11.84LKK109 pKa = 11.03YY110 pKa = 10.2LVPGCAAASLPLLTPFVLPLLLLPLLLVSQTIHH143 pKa = 6.85LSSAQLLVEE152 pKa = 4.73LPFPSPANRR161 pKa = 11.84LFFPCLNPSQSSHH174 pKa = 5.85EE175 pKa = 4.47SVDD178 pKa = 3.73SGCCDD183 pKa = 3.77AEE185 pKa = 4.7GSLNTCSKK193 pKa = 10.53VFNTFSLINTPSQQQQQAPFPSHH216 pKa = 5.24STPPKK221 pKa = 10.14RR222 pKa = 11.84PPLTKK227 pKa = 9.79PRR229 pKa = 11.84SSMKK233 pKa = 10.05KK234 pKa = 10.32DD235 pKa = 3.08EE236 pKa = 4.53TTTPTAKK243 pKa = 10.43AAEE246 pKa = 4.19TAQTGGGASYY256 pKa = 10.76KK257 pKa = 10.14YY258 pKa = 9.31LTWRR262 pKa = 11.84EE263 pKa = 3.08KK264 pKa = 10.4DD265 pKa = 2.74RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84FRR271 pKa = 11.84EE272 pKa = 3.65EE273 pKa = 3.79WKK275 pKa = 10.55HH276 pKa = 5.03LWLVVPHH283 pKa = 6.32GMYY286 pKa = 10.75EE287 pKa = 4.2VMCLVCHH294 pKa = 7.52KK295 pKa = 10.76IMTQRR300 pKa = 11.84KK301 pKa = 7.9LDD303 pKa = 3.76TIKK306 pKa = 10.01RR307 pKa = 11.84HH308 pKa = 5.2TIRR311 pKa = 11.84RR312 pKa = 11.84HH313 pKa = 5.35AEE315 pKa = 3.88LLSMSEE321 pKa = 4.22TEE323 pKa = 4.24RR324 pKa = 11.84QRR326 pKa = 11.84LYY328 pKa = 11.03HH329 pKa = 7.05DD330 pKa = 5.3LVTQYY335 pKa = 10.16FRR337 pKa = 11.84RR338 pKa = 11.84GGDD341 pKa = 3.28FCGEE345 pKa = 4.0SDD347 pKa = 4.16PFCSTSVPRR356 pKa = 11.84LRR358 pKa = 11.84QTGTRR363 pKa = 11.84GSRR366 pKa = 11.84MEE368 pKa = 3.86VAGVRR373 pKa = 11.84RR374 pKa = 11.84HH375 pKa = 6.05SRR377 pKa = 11.84LTDD380 pKa = 3.2VPSPFRR386 pKa = 11.84ARR388 pKa = 11.84MSAPRR393 pKa = 11.84RR394 pKa = 11.84SEE396 pKa = 3.84IDD398 pKa = 3.2THH400 pKa = 6.65PLASSPPSVQTSMSLLPPPPPPPPLPPPLLPTQSDD435 pKa = 3.79TAALLLNRR443 pKa = 4.92
MM1 pKa = 7.45ALHH4 pKa = 6.78RR5 pKa = 11.84RR6 pKa = 11.84LHH8 pKa = 6.06PQPQPPMRR16 pKa = 11.84SPPFRR21 pKa = 11.84PSRR24 pKa = 11.84LPPSTFPSALDD35 pKa = 3.56LTLSLALHH43 pKa = 6.53TCTHH47 pKa = 6.76AGRR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84SRR55 pKa = 11.84SHH57 pKa = 4.92NTPRR61 pKa = 11.84LAPFSQHH68 pKa = 5.7VLLLASRR75 pKa = 11.84SLSCAYY81 pKa = 9.77CQQAVATSSQWLSARR96 pKa = 11.84VSAVRR101 pKa = 11.84SCALGRR107 pKa = 11.84LKK109 pKa = 11.03YY110 pKa = 10.2LVPGCAAASLPLLTPFVLPLLLLPLLLVSQTIHH143 pKa = 6.85LSSAQLLVEE152 pKa = 4.73LPFPSPANRR161 pKa = 11.84LFFPCLNPSQSSHH174 pKa = 5.85EE175 pKa = 4.47SVDD178 pKa = 3.73SGCCDD183 pKa = 3.77AEE185 pKa = 4.7GSLNTCSKK193 pKa = 10.53VFNTFSLINTPSQQQQQAPFPSHH216 pKa = 5.24STPPKK221 pKa = 10.14RR222 pKa = 11.84PPLTKK227 pKa = 9.79PRR229 pKa = 11.84SSMKK233 pKa = 10.05KK234 pKa = 10.32DD235 pKa = 3.08EE236 pKa = 4.53TTTPTAKK243 pKa = 10.43AAEE246 pKa = 4.19TAQTGGGASYY256 pKa = 10.76KK257 pKa = 10.14YY258 pKa = 9.31LTWRR262 pKa = 11.84EE263 pKa = 3.08KK264 pKa = 10.4DD265 pKa = 2.74RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84FRR271 pKa = 11.84EE272 pKa = 3.65EE273 pKa = 3.79WKK275 pKa = 10.55HH276 pKa = 5.03LWLVVPHH283 pKa = 6.32GMYY286 pKa = 10.75EE287 pKa = 4.2VMCLVCHH294 pKa = 7.52KK295 pKa = 10.76IMTQRR300 pKa = 11.84KK301 pKa = 7.9LDD303 pKa = 3.76TIKK306 pKa = 10.01RR307 pKa = 11.84HH308 pKa = 5.2TIRR311 pKa = 11.84RR312 pKa = 11.84HH313 pKa = 5.35AEE315 pKa = 3.88LLSMSEE321 pKa = 4.22TEE323 pKa = 4.24RR324 pKa = 11.84QRR326 pKa = 11.84LYY328 pKa = 11.03HH329 pKa = 7.05DD330 pKa = 5.3LVTQYY335 pKa = 10.16FRR337 pKa = 11.84RR338 pKa = 11.84GGDD341 pKa = 3.28FCGEE345 pKa = 4.0SDD347 pKa = 4.16PFCSTSVPRR356 pKa = 11.84LRR358 pKa = 11.84QTGTRR363 pKa = 11.84GSRR366 pKa = 11.84MEE368 pKa = 3.86VAGVRR373 pKa = 11.84RR374 pKa = 11.84HH375 pKa = 6.05SRR377 pKa = 11.84LTDD380 pKa = 3.2VPSPFRR386 pKa = 11.84ARR388 pKa = 11.84MSAPRR393 pKa = 11.84RR394 pKa = 11.84SEE396 pKa = 3.84IDD398 pKa = 3.2THH400 pKa = 6.65PLASSPPSVQTSMSLLPPPPPPPPLPPPLLPTQSDD435 pKa = 3.79TAALLLNRR443 pKa = 4.92
Molecular weight: 49.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5264045 |
30 |
7853 |
466.7 |
52.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.378 ± 0.023 | 2.203 ± 0.015 |
4.909 ± 0.016 | 6.099 ± 0.027 |
4.087 ± 0.018 | 5.736 ± 0.029 |
2.548 ± 0.011 | 4.977 ± 0.015 |
4.891 ± 0.022 | 9.893 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.142 ± 0.01 | 4.063 ± 0.013 |
5.703 ± 0.026 | 3.98 ± 0.017 |
6.217 ± 0.018 | 9.09 ± 0.028 |
5.912 ± 0.016 | 6.379 ± 0.017 |
1.112 ± 0.008 | 2.679 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
