
Thermomonas sp. SY21
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Thermomonas; unclassified Thermomonas
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
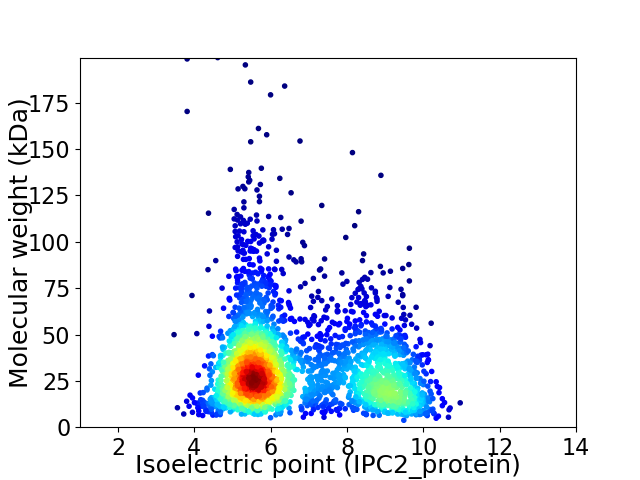
Virtual 2D-PAGE plot for 2753 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
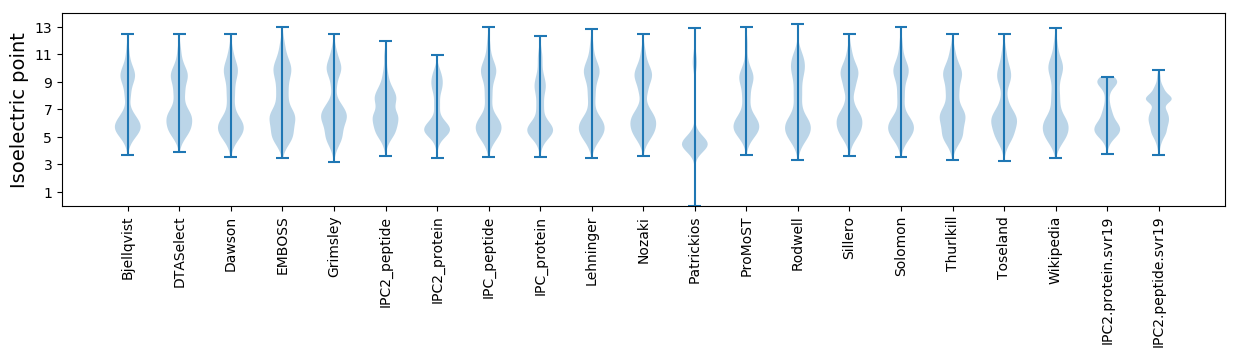
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B7ZTF1|A0A5B7ZTF1_9GAMM Uncharacterized protein OS=Thermomonas sp. SY21 OX=2202149 GN=FHQ07_06950 PE=4 SV=1
MM1 pKa = 7.65SDD3 pKa = 3.12SAAVDD8 pKa = 3.36APYY11 pKa = 8.83RR12 pKa = 11.84TWMCVVCGFVYY23 pKa = 10.68DD24 pKa = 4.02EE25 pKa = 4.77ALGLPEE31 pKa = 6.22DD32 pKa = 4.81GIEE35 pKa = 4.57PGTRR39 pKa = 11.84WEE41 pKa = 5.53DD42 pKa = 3.72IPDD45 pKa = 3.14TWTCPDD51 pKa = 4.14CGVSKK56 pKa = 11.13DD57 pKa = 3.95DD58 pKa = 3.8FEE60 pKa = 5.87LMRR63 pKa = 11.84GG64 pKa = 3.51
MM1 pKa = 7.65SDD3 pKa = 3.12SAAVDD8 pKa = 3.36APYY11 pKa = 8.83RR12 pKa = 11.84TWMCVVCGFVYY23 pKa = 10.68DD24 pKa = 4.02EE25 pKa = 4.77ALGLPEE31 pKa = 6.22DD32 pKa = 4.81GIEE35 pKa = 4.57PGTRR39 pKa = 11.84WEE41 pKa = 5.53DD42 pKa = 3.72IPDD45 pKa = 3.14TWTCPDD51 pKa = 4.14CGVSKK56 pKa = 11.13DD57 pKa = 3.95DD58 pKa = 3.8FEE60 pKa = 5.87LMRR63 pKa = 11.84GG64 pKa = 3.51
Molecular weight: 7.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B7ZNZ4|A0A5B7ZNZ4_9GAMM NADH-quinone oxidoreductase OS=Thermomonas sp. SY21 OX=2202149 GN=nuoG PE=3 SV=1
MM1 pKa = 7.15STGTCAMRR9 pKa = 11.84LPGRR13 pKa = 11.84STNSAISTTPRR24 pKa = 11.84TCAPPISRR32 pKa = 11.84DD33 pKa = 2.98WKK35 pKa = 9.8RR36 pKa = 11.84PPGKK40 pKa = 9.36RR41 pKa = 11.84ARR43 pKa = 11.84RR44 pKa = 11.84AWRR47 pKa = 11.84MPACCRR53 pKa = 11.84PRR55 pKa = 11.84WRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GRR61 pKa = 11.84DD62 pKa = 2.85RR63 pKa = 11.84AAFPLGGRR71 pKa = 11.84PGSRR75 pKa = 11.84RR76 pKa = 11.84LAASWPPRR84 pKa = 11.84VRR86 pKa = 11.84LGVGLGYY93 pKa = 7.78GTCAGPMEE101 pKa = 4.75SAMHH105 pKa = 6.27APNRR109 pKa = 11.84FRR111 pKa = 11.84IALLLCGGALAAGGAMAARR130 pKa = 11.84PAASAGEE137 pKa = 4.34CFASAFEE144 pKa = 4.88SGDD147 pKa = 3.15ADD149 pKa = 5.44AIAACYY155 pKa = 10.6AEE157 pKa = 5.63DD158 pKa = 4.97AIIWFPGGSMAKK170 pKa = 9.78GRR172 pKa = 11.84AAIRR176 pKa = 11.84DD177 pKa = 3.91GFAHH181 pKa = 6.73FLAGATVKK189 pKa = 10.44DD190 pKa = 4.0FQMTNIGQEE199 pKa = 4.21SVGDD203 pKa = 3.92SRR205 pKa = 11.84VSWGTYY211 pKa = 7.27VIRR214 pKa = 11.84MVDD217 pKa = 3.11KK218 pKa = 10.73ASQAEE223 pKa = 4.36SVQTGRR229 pKa = 11.84YY230 pKa = 7.43TDD232 pKa = 3.45VQKK235 pKa = 11.13LVDD238 pKa = 4.23GRR240 pKa = 11.84WLYY243 pKa = 10.56IVDD246 pKa = 4.71HH247 pKa = 7.09PSDD250 pKa = 4.73DD251 pKa = 3.87PAPPAKK257 pKa = 10.44
MM1 pKa = 7.15STGTCAMRR9 pKa = 11.84LPGRR13 pKa = 11.84STNSAISTTPRR24 pKa = 11.84TCAPPISRR32 pKa = 11.84DD33 pKa = 2.98WKK35 pKa = 9.8RR36 pKa = 11.84PPGKK40 pKa = 9.36RR41 pKa = 11.84ARR43 pKa = 11.84RR44 pKa = 11.84AWRR47 pKa = 11.84MPACCRR53 pKa = 11.84PRR55 pKa = 11.84WRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GRR61 pKa = 11.84DD62 pKa = 2.85RR63 pKa = 11.84AAFPLGGRR71 pKa = 11.84PGSRR75 pKa = 11.84RR76 pKa = 11.84LAASWPPRR84 pKa = 11.84VRR86 pKa = 11.84LGVGLGYY93 pKa = 7.78GTCAGPMEE101 pKa = 4.75SAMHH105 pKa = 6.27APNRR109 pKa = 11.84FRR111 pKa = 11.84IALLLCGGALAAGGAMAARR130 pKa = 11.84PAASAGEE137 pKa = 4.34CFASAFEE144 pKa = 4.88SGDD147 pKa = 3.15ADD149 pKa = 5.44AIAACYY155 pKa = 10.6AEE157 pKa = 5.63DD158 pKa = 4.97AIIWFPGGSMAKK170 pKa = 9.78GRR172 pKa = 11.84AAIRR176 pKa = 11.84DD177 pKa = 3.91GFAHH181 pKa = 6.73FLAGATVKK189 pKa = 10.44DD190 pKa = 4.0FQMTNIGQEE199 pKa = 4.21SVGDD203 pKa = 3.92SRR205 pKa = 11.84VSWGTYY211 pKa = 7.27VIRR214 pKa = 11.84MVDD217 pKa = 3.11KK218 pKa = 10.73ASQAEE223 pKa = 4.36SVQTGRR229 pKa = 11.84YY230 pKa = 7.43TDD232 pKa = 3.45VQKK235 pKa = 11.13LVDD238 pKa = 4.23GRR240 pKa = 11.84WLYY243 pKa = 10.56IVDD246 pKa = 4.71HH247 pKa = 7.09PSDD250 pKa = 4.73DD251 pKa = 3.87PAPPAKK257 pKa = 10.44
Molecular weight: 27.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
912151 |
32 |
2007 |
331.3 |
35.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.764 ± 0.083 | 0.86 ± 0.016 |
6.076 ± 0.036 | 5.268 ± 0.042 |
3.412 ± 0.03 | 8.761 ± 0.053 |
2.188 ± 0.025 | 4.215 ± 0.03 |
3.173 ± 0.04 | 10.745 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.261 ± 0.02 | 2.529 ± 0.034 |
5.211 ± 0.038 | 3.625 ± 0.028 |
7.698 ± 0.054 | 4.894 ± 0.037 |
4.422 ± 0.042 | 7.041 ± 0.038 |
1.533 ± 0.021 | 2.324 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
