
Phenylobacterium sp. Root1277
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Phenylobacterium; unclassified Phenylobacterium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
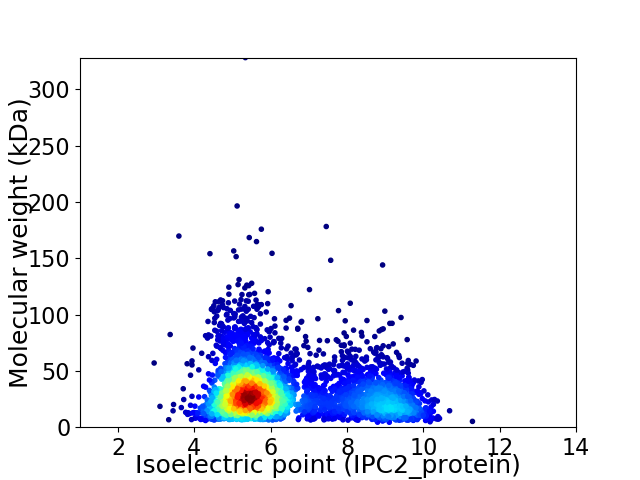
Virtual 2D-PAGE plot for 4022 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7D775|A0A0Q7D775_9CAUL Uncharacterized protein OS=Phenylobacterium sp. Root1277 OX=1736442 GN=ASC73_17545 PE=4 SV=1
MM1 pKa = 7.17FVDD4 pKa = 3.81TRR6 pKa = 11.84VEE8 pKa = 4.25GYY10 pKa = 7.24EE11 pKa = 4.1TIVTGVAPGAEE22 pKa = 4.19VVLIDD27 pKa = 4.56ASGDD31 pKa = 3.3GVAQMAQALAGRR43 pKa = 11.84TDD45 pKa = 2.72ITAIHH50 pKa = 6.37IVGHH54 pKa = 5.26GQAGVMMLGSATLGAGDD71 pKa = 3.33IDD73 pKa = 4.4AYY75 pKa = 10.65AGEE78 pKa = 4.15LRR80 pKa = 11.84AIGGALAPDD89 pKa = 4.49GDD91 pKa = 3.92IQLWGCEE98 pKa = 3.73IGAGPEE104 pKa = 3.64GAAFIQALAAATGADD119 pKa = 3.2IAASDD124 pKa = 4.53DD125 pKa = 3.6LTGAGGDD132 pKa = 3.49WVLEE136 pKa = 4.37TQTGTIEE143 pKa = 4.04TSVAITPAGQHH154 pKa = 5.82AFAGTLAIGSEE165 pKa = 4.26NFDD168 pKa = 3.61SLGLITTFGTTLTAGNWQFTSGSAADD194 pKa = 3.88HH195 pKa = 6.36AVASSPEE202 pKa = 3.86YY203 pKa = 10.76AVWLNNDD210 pKa = 3.09GGASDD215 pKa = 3.88RR216 pKa = 11.84AVILNYY222 pKa = 10.8NGDD225 pKa = 4.03NITNYY230 pKa = 10.76AMKK233 pKa = 10.61SSDD236 pKa = 3.18GTNFALQSFKK246 pKa = 10.53IGQSAGATQTLTIAAYY262 pKa = 9.8SNGVQVVAPEE272 pKa = 4.32TVNLAASDD280 pKa = 4.1SIGGVNYY287 pKa = 10.79ALGGNDD293 pKa = 3.15GSGAYY298 pKa = 7.58GTLTFSSAYY307 pKa = 10.66ANIDD311 pKa = 3.32EE312 pKa = 4.3IRR314 pKa = 11.84FVFGGLSDD322 pKa = 4.31LHH324 pKa = 8.4IDD326 pKa = 4.87DD327 pKa = 4.48IAVAPAVVDD336 pKa = 3.38VTAPVFVSAAVNGATLVMTYY356 pKa = 10.78NEE358 pKa = 5.7AITLDD363 pKa = 3.81AVNAPAAGRR372 pKa = 11.84FAVMADD378 pKa = 3.57GASVTVNSVTVNAAAKK394 pKa = 8.86TVTLTLQSAVTAGQAVTVAYY414 pKa = 8.05TDD416 pKa = 3.52PTGGDD421 pKa = 3.59DD422 pKa = 4.74ASAIQDD428 pKa = 3.51AVGNDD433 pKa = 3.72AASLPATSVTNNTVDD448 pKa = 3.74PPPLVSGNIAVPADD462 pKa = 3.35GSYY465 pKa = 11.08AAGQTLSFTVTFTEE479 pKa = 4.06NVTVVGTDD487 pKa = 3.09SVLGLTIGSTARR499 pKa = 11.84EE500 pKa = 3.92AVFASSTANSVTYY513 pKa = 10.14AYY515 pKa = 8.44TVQAGDD521 pKa = 3.27TDD523 pKa = 4.49ANGVAVGALNLNTSTIRR540 pKa = 11.84DD541 pKa = 3.69VAGNDD546 pKa = 3.13ANLALGGHH554 pKa = 5.78VPSTAGVLVDD564 pKa = 3.37TTVPEE569 pKa = 4.24VATVSVPSGGTYY581 pKa = 10.63AAGQTLSFTVNFDD594 pKa = 4.21DD595 pKa = 3.98NVTVAGTDD603 pKa = 3.2STLALTIGATARR615 pKa = 11.84TATYY619 pKa = 10.49LSKK622 pKa = 9.49TANSITYY629 pKa = 8.48TYY631 pKa = 9.39TVQAGEE637 pKa = 4.11TDD639 pKa = 3.15ADD641 pKa = 4.81GITVGALSKK650 pKa = 10.8GSSTIRR656 pKa = 11.84DD657 pKa = 3.4AAGNDD662 pKa = 3.27AVLTLNSVGATSGVLVDD679 pKa = 3.55ATVPTVSGNISVPTGGMYY697 pKa = 9.46MAGQTLSFTVTFDD710 pKa = 3.58EE711 pKa = 4.31NVTLSGSPSTLGLTIGAVSRR731 pKa = 11.84SADD734 pKa = 3.18ITGTTGNSITYY745 pKa = 9.37SYY747 pKa = 9.82TVQASDD753 pKa = 3.53YY754 pKa = 9.32DD755 pKa = 3.95ADD757 pKa = 4.83GIAVTGISLNGGTIRR772 pKa = 11.84DD773 pKa = 3.86GAGNNADD780 pKa = 3.82LTLSGHH786 pKa = 6.48LPSLAGVLVDD796 pKa = 4.37GGPPAVSGNIAVPANGSYY814 pKa = 11.39GEE816 pKa = 4.4GATLTFTVTFGEE828 pKa = 4.41NVTVTGSDD836 pKa = 3.13SVLGLTIGSIARR848 pKa = 11.84EE849 pKa = 3.91ATYY852 pKa = 10.71ASSSGASVTYY862 pKa = 10.14AYY864 pKa = 8.54TVQPGEE870 pKa = 3.91TDD872 pKa = 3.12ANGIAIGAVNLNTSTIKK889 pKa = 10.78DD890 pKa = 3.33AAGNHH895 pKa = 6.24ANLALGGHH903 pKa = 6.08VPATSGILVDD913 pKa = 3.89TTAPGVASVSVPSSATYY930 pKa = 10.57VAGQNLDD937 pKa = 4.01FIVNFDD943 pKa = 3.82EE944 pKa = 4.56AVTIAGSDD952 pKa = 3.56SVIALMVGSTSRR964 pKa = 11.84SATFFSSSGSTATYY978 pKa = 10.0RR979 pKa = 11.84YY980 pKa = 7.06TVQAGDD986 pKa = 3.32VDD988 pKa = 4.08ADD990 pKa = 4.48GITVGALSKK999 pKa = 10.8GSSTIRR1005 pKa = 11.84DD1006 pKa = 3.54SAGNDD1011 pKa = 3.15AEE1013 pKa = 4.39LTLNSVGATNGVMIDD1028 pKa = 3.51GTVPSVTGNVSVPANDD1044 pKa = 3.64TYY1046 pKa = 11.81VVGEE1050 pKa = 4.44TPSFTVTFDD1059 pKa = 3.42EE1060 pKa = 4.41NVTVTGTGSTLGLTIGATARR1080 pKa = 11.84TAAYY1084 pKa = 9.98ASKK1087 pKa = 8.46TANSITYY1094 pKa = 8.48TYY1096 pKa = 9.82TVQAGDD1102 pKa = 3.61NDD1104 pKa = 3.9ADD1106 pKa = 4.25GIVVNGISLNGGTIRR1121 pKa = 11.84DD1122 pKa = 3.81GAGNNANLSLTGHH1135 pKa = 6.73IPATTGVLVDD1145 pKa = 3.9TAAPSFTSATVNGATLVMTYY1165 pKa = 10.68SDD1167 pKa = 5.24AGLLDD1172 pKa = 4.67AGHH1175 pKa = 7.13APGAGAFTVMVGGAPVSVTAVAVNGAAHH1203 pKa = 5.87TVSLTLASPVAAGAAVTVAYY1223 pKa = 10.43VDD1225 pKa = 3.75PTAGDD1230 pKa = 3.87DD1231 pKa = 3.52VSAIQDD1237 pKa = 3.35AGRR1240 pKa = 11.84NDD1242 pKa = 3.97AASLAATSVTNTTAAPPTTPPTTPPVTTTVDD1273 pKa = 3.54GVPVQTQTSTASDD1286 pKa = 3.69GSVSQIVTVLVVQPGRR1302 pKa = 11.84VEE1304 pKa = 3.8QVGNNTVADD1313 pKa = 3.73IPLVKK1318 pKa = 10.58DD1319 pKa = 3.54GAGQALLALQVPVGYY1334 pKa = 10.49GLQASGAAQPKK1345 pKa = 9.58AAGDD1349 pKa = 3.83ALSDD1353 pKa = 4.37LIRR1356 pKa = 11.84EE1357 pKa = 4.32IRR1359 pKa = 11.84AHH1361 pKa = 5.96TNSSSQDD1368 pKa = 3.18QGQLGGAGTDD1378 pKa = 3.86FVNGLGASTPLLVQTIVTTGADD1400 pKa = 3.91AAPGAQPLTISGQPQAQGQPQTALVIDD1427 pKa = 4.34GRR1429 pKa = 11.84SNPGIHH1435 pKa = 7.08IEE1437 pKa = 4.04LQNVEE1442 pKa = 4.25FAAIIGSATVTGGAGSQHH1460 pKa = 5.14VWGDD1464 pKa = 3.54GASQTIFLGADD1475 pKa = 3.32DD1476 pKa = 5.41DD1477 pKa = 4.36VLHH1480 pKa = 6.94GGGGNDD1486 pKa = 3.44TVGSAGGNDD1495 pKa = 3.56EE1496 pKa = 5.13VYY1498 pKa = 10.99GDD1500 pKa = 4.78DD1501 pKa = 4.01GDD1503 pKa = 4.27DD1504 pKa = 3.49VVVGGEE1510 pKa = 4.08GADD1513 pKa = 3.61FVHH1516 pKa = 6.95GNAGNDD1522 pKa = 3.63TARR1525 pKa = 11.84GDD1527 pKa = 3.66AGDD1530 pKa = 3.93DD1531 pKa = 3.37LVYY1534 pKa = 10.69GGKK1537 pKa = 10.53GDD1539 pKa = 3.78DD1540 pKa = 3.69QVFGGADD1547 pKa = 3.49NDD1549 pKa = 4.65LLFGDD1554 pKa = 5.1LGQDD1558 pKa = 3.33TLQGNTGHH1566 pKa = 6.97DD1567 pKa = 3.87TLDD1570 pKa = 3.73GGADD1574 pKa = 3.53NDD1576 pKa = 5.23LMFGGQNNDD1585 pKa = 3.42LLLGGEE1591 pKa = 4.35GDD1593 pKa = 4.27DD1594 pKa = 5.16RR1595 pKa = 11.84LNGDD1599 pKa = 4.76LGNDD1603 pKa = 3.63TLQGNVGRR1611 pKa = 11.84DD1612 pKa = 3.56TLEE1615 pKa = 4.31GGAGADD1621 pKa = 3.78FLYY1624 pKa = 10.64GGQDD1628 pKa = 3.18NDD1630 pKa = 3.64VLSGGEE1636 pKa = 4.08GADD1639 pKa = 3.28TLFGDD1644 pKa = 4.86RR1645 pKa = 11.84GDD1647 pKa = 3.76DD1648 pKa = 3.67TLIGGAGADD1657 pKa = 3.68LFVTASGRR1665 pKa = 11.84GEE1667 pKa = 4.13DD1668 pKa = 3.91QVLDD1672 pKa = 4.55FAAGDD1677 pKa = 3.85RR1678 pKa = 11.84VGLLAGTTYY1687 pKa = 10.92SAAQVGGDD1695 pKa = 3.55TVITLTGGGQTTLVNVQLSGLGDD1718 pKa = 3.37GWITTII1724 pKa = 4.94
MM1 pKa = 7.17FVDD4 pKa = 3.81TRR6 pKa = 11.84VEE8 pKa = 4.25GYY10 pKa = 7.24EE11 pKa = 4.1TIVTGVAPGAEE22 pKa = 4.19VVLIDD27 pKa = 4.56ASGDD31 pKa = 3.3GVAQMAQALAGRR43 pKa = 11.84TDD45 pKa = 2.72ITAIHH50 pKa = 6.37IVGHH54 pKa = 5.26GQAGVMMLGSATLGAGDD71 pKa = 3.33IDD73 pKa = 4.4AYY75 pKa = 10.65AGEE78 pKa = 4.15LRR80 pKa = 11.84AIGGALAPDD89 pKa = 4.49GDD91 pKa = 3.92IQLWGCEE98 pKa = 3.73IGAGPEE104 pKa = 3.64GAAFIQALAAATGADD119 pKa = 3.2IAASDD124 pKa = 4.53DD125 pKa = 3.6LTGAGGDD132 pKa = 3.49WVLEE136 pKa = 4.37TQTGTIEE143 pKa = 4.04TSVAITPAGQHH154 pKa = 5.82AFAGTLAIGSEE165 pKa = 4.26NFDD168 pKa = 3.61SLGLITTFGTTLTAGNWQFTSGSAADD194 pKa = 3.88HH195 pKa = 6.36AVASSPEE202 pKa = 3.86YY203 pKa = 10.76AVWLNNDD210 pKa = 3.09GGASDD215 pKa = 3.88RR216 pKa = 11.84AVILNYY222 pKa = 10.8NGDD225 pKa = 4.03NITNYY230 pKa = 10.76AMKK233 pKa = 10.61SSDD236 pKa = 3.18GTNFALQSFKK246 pKa = 10.53IGQSAGATQTLTIAAYY262 pKa = 9.8SNGVQVVAPEE272 pKa = 4.32TVNLAASDD280 pKa = 4.1SIGGVNYY287 pKa = 10.79ALGGNDD293 pKa = 3.15GSGAYY298 pKa = 7.58GTLTFSSAYY307 pKa = 10.66ANIDD311 pKa = 3.32EE312 pKa = 4.3IRR314 pKa = 11.84FVFGGLSDD322 pKa = 4.31LHH324 pKa = 8.4IDD326 pKa = 4.87DD327 pKa = 4.48IAVAPAVVDD336 pKa = 3.38VTAPVFVSAAVNGATLVMTYY356 pKa = 10.78NEE358 pKa = 5.7AITLDD363 pKa = 3.81AVNAPAAGRR372 pKa = 11.84FAVMADD378 pKa = 3.57GASVTVNSVTVNAAAKK394 pKa = 8.86TVTLTLQSAVTAGQAVTVAYY414 pKa = 8.05TDD416 pKa = 3.52PTGGDD421 pKa = 3.59DD422 pKa = 4.74ASAIQDD428 pKa = 3.51AVGNDD433 pKa = 3.72AASLPATSVTNNTVDD448 pKa = 3.74PPPLVSGNIAVPADD462 pKa = 3.35GSYY465 pKa = 11.08AAGQTLSFTVTFTEE479 pKa = 4.06NVTVVGTDD487 pKa = 3.09SVLGLTIGSTARR499 pKa = 11.84EE500 pKa = 3.92AVFASSTANSVTYY513 pKa = 10.14AYY515 pKa = 8.44TVQAGDD521 pKa = 3.27TDD523 pKa = 4.49ANGVAVGALNLNTSTIRR540 pKa = 11.84DD541 pKa = 3.69VAGNDD546 pKa = 3.13ANLALGGHH554 pKa = 5.78VPSTAGVLVDD564 pKa = 3.37TTVPEE569 pKa = 4.24VATVSVPSGGTYY581 pKa = 10.63AAGQTLSFTVNFDD594 pKa = 4.21DD595 pKa = 3.98NVTVAGTDD603 pKa = 3.2STLALTIGATARR615 pKa = 11.84TATYY619 pKa = 10.49LSKK622 pKa = 9.49TANSITYY629 pKa = 8.48TYY631 pKa = 9.39TVQAGEE637 pKa = 4.11TDD639 pKa = 3.15ADD641 pKa = 4.81GITVGALSKK650 pKa = 10.8GSSTIRR656 pKa = 11.84DD657 pKa = 3.4AAGNDD662 pKa = 3.27AVLTLNSVGATSGVLVDD679 pKa = 3.55ATVPTVSGNISVPTGGMYY697 pKa = 9.46MAGQTLSFTVTFDD710 pKa = 3.58EE711 pKa = 4.31NVTLSGSPSTLGLTIGAVSRR731 pKa = 11.84SADD734 pKa = 3.18ITGTTGNSITYY745 pKa = 9.37SYY747 pKa = 9.82TVQASDD753 pKa = 3.53YY754 pKa = 9.32DD755 pKa = 3.95ADD757 pKa = 4.83GIAVTGISLNGGTIRR772 pKa = 11.84DD773 pKa = 3.86GAGNNADD780 pKa = 3.82LTLSGHH786 pKa = 6.48LPSLAGVLVDD796 pKa = 4.37GGPPAVSGNIAVPANGSYY814 pKa = 11.39GEE816 pKa = 4.4GATLTFTVTFGEE828 pKa = 4.41NVTVTGSDD836 pKa = 3.13SVLGLTIGSIARR848 pKa = 11.84EE849 pKa = 3.91ATYY852 pKa = 10.71ASSSGASVTYY862 pKa = 10.14AYY864 pKa = 8.54TVQPGEE870 pKa = 3.91TDD872 pKa = 3.12ANGIAIGAVNLNTSTIKK889 pKa = 10.78DD890 pKa = 3.33AAGNHH895 pKa = 6.24ANLALGGHH903 pKa = 6.08VPATSGILVDD913 pKa = 3.89TTAPGVASVSVPSSATYY930 pKa = 10.57VAGQNLDD937 pKa = 4.01FIVNFDD943 pKa = 3.82EE944 pKa = 4.56AVTIAGSDD952 pKa = 3.56SVIALMVGSTSRR964 pKa = 11.84SATFFSSSGSTATYY978 pKa = 10.0RR979 pKa = 11.84YY980 pKa = 7.06TVQAGDD986 pKa = 3.32VDD988 pKa = 4.08ADD990 pKa = 4.48GITVGALSKK999 pKa = 10.8GSSTIRR1005 pKa = 11.84DD1006 pKa = 3.54SAGNDD1011 pKa = 3.15AEE1013 pKa = 4.39LTLNSVGATNGVMIDD1028 pKa = 3.51GTVPSVTGNVSVPANDD1044 pKa = 3.64TYY1046 pKa = 11.81VVGEE1050 pKa = 4.44TPSFTVTFDD1059 pKa = 3.42EE1060 pKa = 4.41NVTVTGTGSTLGLTIGATARR1080 pKa = 11.84TAAYY1084 pKa = 9.98ASKK1087 pKa = 8.46TANSITYY1094 pKa = 8.48TYY1096 pKa = 9.82TVQAGDD1102 pKa = 3.61NDD1104 pKa = 3.9ADD1106 pKa = 4.25GIVVNGISLNGGTIRR1121 pKa = 11.84DD1122 pKa = 3.81GAGNNANLSLTGHH1135 pKa = 6.73IPATTGVLVDD1145 pKa = 3.9TAAPSFTSATVNGATLVMTYY1165 pKa = 10.68SDD1167 pKa = 5.24AGLLDD1172 pKa = 4.67AGHH1175 pKa = 7.13APGAGAFTVMVGGAPVSVTAVAVNGAAHH1203 pKa = 5.87TVSLTLASPVAAGAAVTVAYY1223 pKa = 10.43VDD1225 pKa = 3.75PTAGDD1230 pKa = 3.87DD1231 pKa = 3.52VSAIQDD1237 pKa = 3.35AGRR1240 pKa = 11.84NDD1242 pKa = 3.97AASLAATSVTNTTAAPPTTPPTTPPVTTTVDD1273 pKa = 3.54GVPVQTQTSTASDD1286 pKa = 3.69GSVSQIVTVLVVQPGRR1302 pKa = 11.84VEE1304 pKa = 3.8QVGNNTVADD1313 pKa = 3.73IPLVKK1318 pKa = 10.58DD1319 pKa = 3.54GAGQALLALQVPVGYY1334 pKa = 10.49GLQASGAAQPKK1345 pKa = 9.58AAGDD1349 pKa = 3.83ALSDD1353 pKa = 4.37LIRR1356 pKa = 11.84EE1357 pKa = 4.32IRR1359 pKa = 11.84AHH1361 pKa = 5.96TNSSSQDD1368 pKa = 3.18QGQLGGAGTDD1378 pKa = 3.86FVNGLGASTPLLVQTIVTTGADD1400 pKa = 3.91AAPGAQPLTISGQPQAQGQPQTALVIDD1427 pKa = 4.34GRR1429 pKa = 11.84SNPGIHH1435 pKa = 7.08IEE1437 pKa = 4.04LQNVEE1442 pKa = 4.25FAAIIGSATVTGGAGSQHH1460 pKa = 5.14VWGDD1464 pKa = 3.54GASQTIFLGADD1475 pKa = 3.32DD1476 pKa = 5.41DD1477 pKa = 4.36VLHH1480 pKa = 6.94GGGGNDD1486 pKa = 3.44TVGSAGGNDD1495 pKa = 3.56EE1496 pKa = 5.13VYY1498 pKa = 10.99GDD1500 pKa = 4.78DD1501 pKa = 4.01GDD1503 pKa = 4.27DD1504 pKa = 3.49VVVGGEE1510 pKa = 4.08GADD1513 pKa = 3.61FVHH1516 pKa = 6.95GNAGNDD1522 pKa = 3.63TARR1525 pKa = 11.84GDD1527 pKa = 3.66AGDD1530 pKa = 3.93DD1531 pKa = 3.37LVYY1534 pKa = 10.69GGKK1537 pKa = 10.53GDD1539 pKa = 3.78DD1540 pKa = 3.69QVFGGADD1547 pKa = 3.49NDD1549 pKa = 4.65LLFGDD1554 pKa = 5.1LGQDD1558 pKa = 3.33TLQGNTGHH1566 pKa = 6.97DD1567 pKa = 3.87TLDD1570 pKa = 3.73GGADD1574 pKa = 3.53NDD1576 pKa = 5.23LMFGGQNNDD1585 pKa = 3.42LLLGGEE1591 pKa = 4.35GDD1593 pKa = 4.27DD1594 pKa = 5.16RR1595 pKa = 11.84LNGDD1599 pKa = 4.76LGNDD1603 pKa = 3.63TLQGNVGRR1611 pKa = 11.84DD1612 pKa = 3.56TLEE1615 pKa = 4.31GGAGADD1621 pKa = 3.78FLYY1624 pKa = 10.64GGQDD1628 pKa = 3.18NDD1630 pKa = 3.64VLSGGEE1636 pKa = 4.08GADD1639 pKa = 3.28TLFGDD1644 pKa = 4.86RR1645 pKa = 11.84GDD1647 pKa = 3.76DD1648 pKa = 3.67TLIGGAGADD1657 pKa = 3.68LFVTASGRR1665 pKa = 11.84GEE1667 pKa = 4.13DD1668 pKa = 3.91QVLDD1672 pKa = 4.55FAAGDD1677 pKa = 3.85RR1678 pKa = 11.84VGLLAGTTYY1687 pKa = 10.92SAAQVGGDD1695 pKa = 3.55TVITLTGGGQTTLVNVQLSGLGDD1718 pKa = 3.37GWITTII1724 pKa = 4.94
Molecular weight: 169.78 kDa
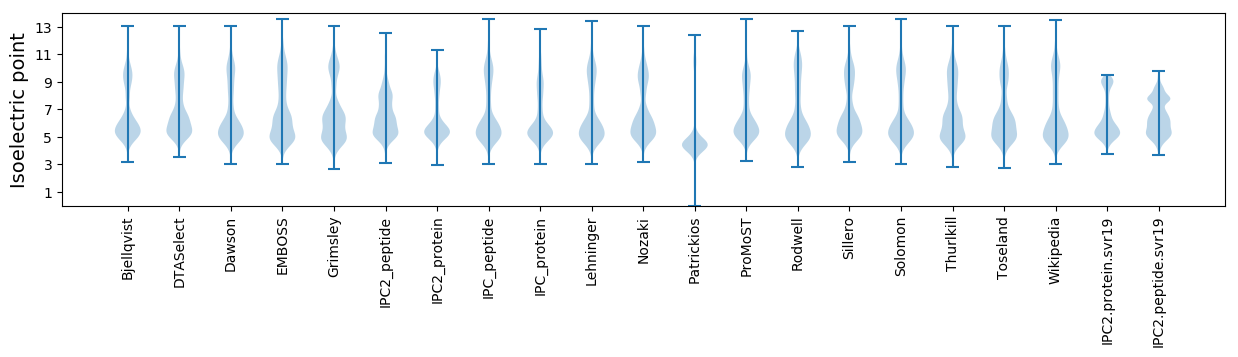
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7D7K4|A0A0Q7D7K4_9CAUL Uncharacterized protein OS=Phenylobacterium sp. Root1277 OX=1736442 GN=ASC73_14245 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGAKK29 pKa = 9.48IVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGAKK29 pKa = 9.48IVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1269598 |
41 |
3085 |
315.7 |
33.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.998 ± 0.055 | 0.731 ± 0.013 |
5.754 ± 0.026 | 5.7 ± 0.038 |
3.626 ± 0.023 | 9.034 ± 0.038 |
1.804 ± 0.019 | 4.411 ± 0.028 |
3.171 ± 0.031 | 9.977 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.342 ± 0.018 | 2.353 ± 0.026 |
5.445 ± 0.034 | 3.23 ± 0.022 |
7.107 ± 0.038 | 5.086 ± 0.024 |
5.076 ± 0.03 | 7.542 ± 0.028 |
1.41 ± 0.017 | 2.203 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
