
Betta splendens (Siamese fighting fish)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia; Ctenosquamata; Acanthomorphata;
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
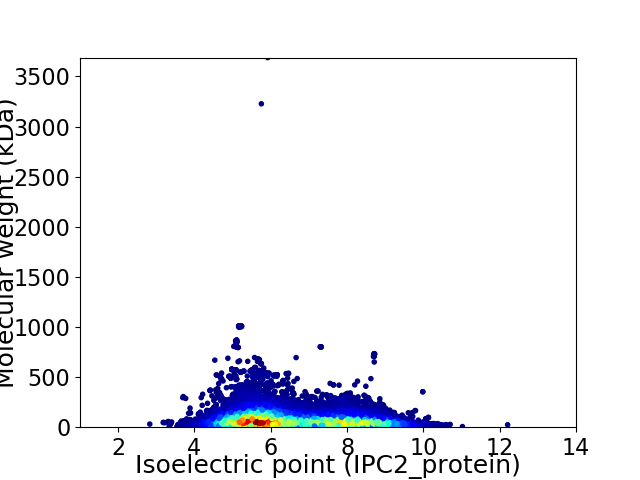
Virtual 2D-PAGE plot for 41292 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P7LUX0|A0A6P7LUX0_BETSP transcription factor E2F6-like OS=Betta splendens OX=158456 GN=LOC114850007 PE=3 SV=1
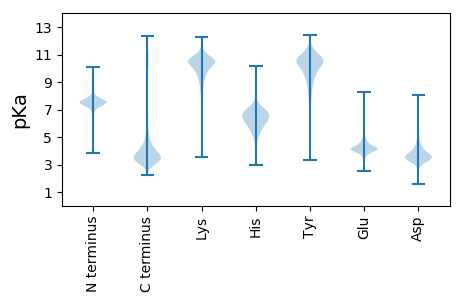
MM1 pKa = 7.9DD2 pKa = 5.47LSFMAAQIPVMTGAFMDD19 pKa = 4.02SSPNDD24 pKa = 3.88DD25 pKa = 3.69YY26 pKa = 11.86STDD29 pKa = 3.18HH30 pKa = 6.97SLFNSSASVHH40 pKa = 5.71AASMVAQSPPEE51 pKa = 4.19EE52 pKa = 4.27PQSMSRR58 pKa = 11.84DD59 pKa = 4.1AIWLWIAITATIGNIVVVGVVYY81 pKa = 10.98AFTFF85 pKa = 3.7
MM1 pKa = 7.9DD2 pKa = 5.47LSFMAAQIPVMTGAFMDD19 pKa = 4.02SSPNDD24 pKa = 3.88DD25 pKa = 3.69YY26 pKa = 11.86STDD29 pKa = 3.18HH30 pKa = 6.97SLFNSSASVHH40 pKa = 5.71AASMVAQSPPEE51 pKa = 4.19EE52 pKa = 4.27PQSMSRR58 pKa = 11.84DD59 pKa = 4.1AIWLWIAITATIGNIVVVGVVYY81 pKa = 10.98AFTFF85 pKa = 3.7
Molecular weight: 9.15 kDa
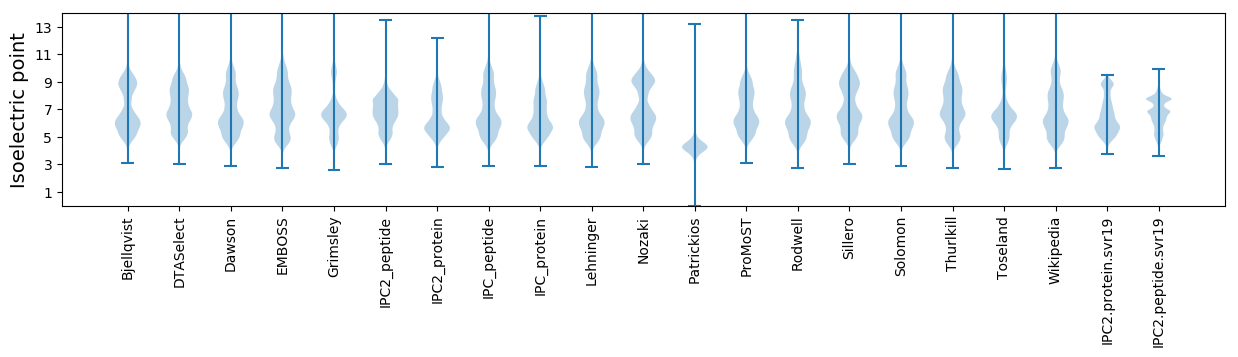
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P7NV97|A0A6P7NV97_BETSP Isoform of A0A6P7P2G4 tuftelin-like isoform X4 OS=Betta splendens OX=158456 GN=LOC114866266 PE=4 SV=1
MM1 pKa = 7.76AFMSAKK7 pKa = 10.31LEE9 pKa = 4.09RR10 pKa = 11.84SGVWSVPAEE19 pKa = 3.78QVSTNRR25 pKa = 11.84SASTGQRR32 pKa = 11.84PQVSTNRR39 pKa = 11.84SASTGQRR46 pKa = 11.84LQVSANRR53 pKa = 11.84STPTGQRR60 pKa = 11.84QQVSANRR67 pKa = 11.84SAPTGQRR74 pKa = 11.84QQVNASRR81 pKa = 11.84SAPTGQRR88 pKa = 11.84PQVSANRR95 pKa = 11.84SAPTGQRR102 pKa = 11.84QQVNASRR109 pKa = 11.84SAPTGQRR116 pKa = 11.84QQVNASRR123 pKa = 11.84SAPTGQRR130 pKa = 11.84QQVSAHH136 pKa = 6.38RR137 pKa = 11.84SASTGQRR144 pKa = 11.84PQVNANRR151 pKa = 11.84STPRR155 pKa = 11.84VNAHH159 pKa = 6.23RR160 pKa = 11.84SVPTGQRR167 pKa = 11.84PQVNAHH173 pKa = 6.75RR174 pKa = 11.84STPTGQRR181 pKa = 11.84PQVNANRR188 pKa = 11.84STPRR192 pKa = 11.84VNAHH196 pKa = 6.23RR197 pKa = 11.84SVPTGQRR204 pKa = 11.84PP205 pKa = 3.04
MM1 pKa = 7.76AFMSAKK7 pKa = 10.31LEE9 pKa = 4.09RR10 pKa = 11.84SGVWSVPAEE19 pKa = 3.78QVSTNRR25 pKa = 11.84SASTGQRR32 pKa = 11.84PQVSTNRR39 pKa = 11.84SASTGQRR46 pKa = 11.84LQVSANRR53 pKa = 11.84STPTGQRR60 pKa = 11.84QQVSANRR67 pKa = 11.84SAPTGQRR74 pKa = 11.84QQVNASRR81 pKa = 11.84SAPTGQRR88 pKa = 11.84PQVSANRR95 pKa = 11.84SAPTGQRR102 pKa = 11.84QQVNASRR109 pKa = 11.84SAPTGQRR116 pKa = 11.84QQVNASRR123 pKa = 11.84SAPTGQRR130 pKa = 11.84QQVSAHH136 pKa = 6.38RR137 pKa = 11.84SASTGQRR144 pKa = 11.84PQVNANRR151 pKa = 11.84STPRR155 pKa = 11.84VNAHH159 pKa = 6.23RR160 pKa = 11.84SVPTGQRR167 pKa = 11.84PQVNAHH173 pKa = 6.75RR174 pKa = 11.84STPTGQRR181 pKa = 11.84PQVNANRR188 pKa = 11.84STPRR192 pKa = 11.84VNAHH196 pKa = 6.23RR197 pKa = 11.84SVPTGQRR204 pKa = 11.84PP205 pKa = 3.04
Molecular weight: 21.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
32944339 |
31 |
33161 |
797.8 |
88.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.935 ± 0.01 | 2.089 ± 0.011 |
5.286 ± 0.008 | 7.023 ± 0.018 |
3.273 ± 0.009 | 6.379 ± 0.016 |
2.663 ± 0.006 | 4.033 ± 0.009 |
5.541 ± 0.016 | 9.378 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.256 ± 0.006 | 3.688 ± 0.007 |
5.97 ± 0.02 | 4.966 ± 0.013 |
5.84 ± 0.009 | 9.064 ± 0.015 |
5.695 ± 0.013 | 6.293 ± 0.011 |
1.079 ± 0.004 | 2.548 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
