
Apis mellifera associated microvirus 35
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
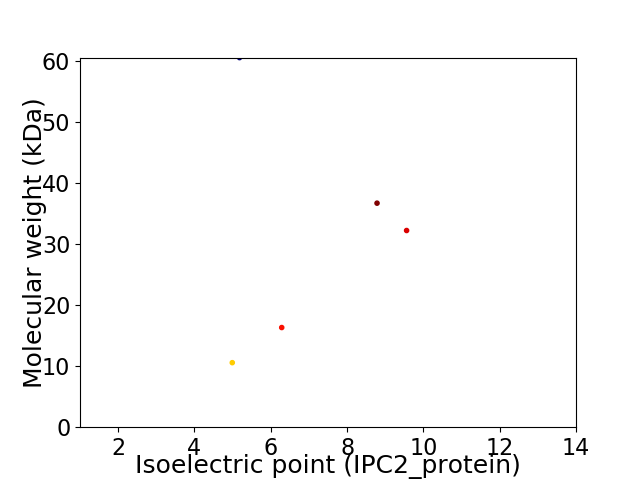
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
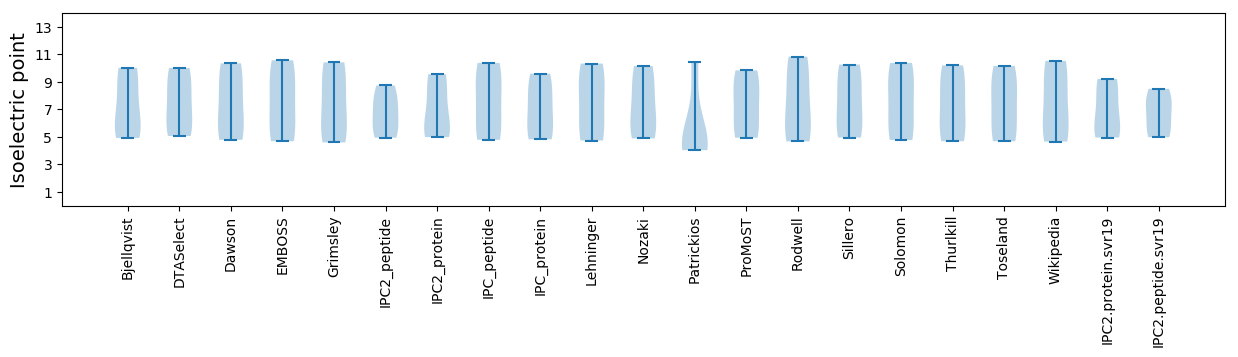
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U5V0|A0A3Q8U5V0_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 35 OX=2494764 PE=3 SV=1
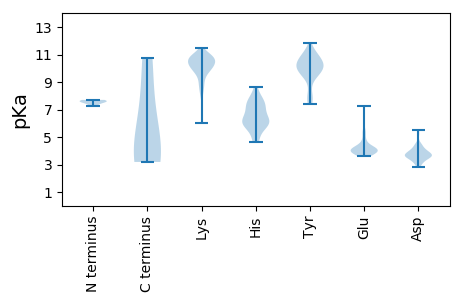
MM1 pKa = 7.64SEE3 pKa = 3.81KK4 pKa = 9.92TKK6 pKa = 10.9QFVGSVKK13 pKa = 10.6AKK15 pKa = 10.02TDD17 pKa = 3.72FAVLNNADD25 pKa = 3.76IDD27 pKa = 3.97RR28 pKa = 11.84DD29 pKa = 3.94QIKK32 pKa = 9.87EE33 pKa = 4.04WLVQDD38 pKa = 3.98VKK40 pKa = 10.98RR41 pKa = 11.84VAILVNDD48 pKa = 3.95ILNTPEE54 pKa = 4.88CIEE57 pKa = 3.99NLTDD61 pKa = 3.41VFYY64 pKa = 11.23KK65 pKa = 10.44RR66 pKa = 11.84YY67 pKa = 9.73QDD69 pKa = 3.38FHH71 pKa = 8.63KK72 pKa = 10.77EE73 pKa = 3.73NAIKK77 pKa = 10.41EE78 pKa = 4.28MAEE81 pKa = 3.79KK82 pKa = 10.36DD83 pKa = 3.81YY84 pKa = 11.22STVEE88 pKa = 3.87QNGG91 pKa = 3.19
MM1 pKa = 7.64SEE3 pKa = 3.81KK4 pKa = 9.92TKK6 pKa = 10.9QFVGSVKK13 pKa = 10.6AKK15 pKa = 10.02TDD17 pKa = 3.72FAVLNNADD25 pKa = 3.76IDD27 pKa = 3.97RR28 pKa = 11.84DD29 pKa = 3.94QIKK32 pKa = 9.87EE33 pKa = 4.04WLVQDD38 pKa = 3.98VKK40 pKa = 10.98RR41 pKa = 11.84VAILVNDD48 pKa = 3.95ILNTPEE54 pKa = 4.88CIEE57 pKa = 3.99NLTDD61 pKa = 3.41VFYY64 pKa = 11.23KK65 pKa = 10.44RR66 pKa = 11.84YY67 pKa = 9.73QDD69 pKa = 3.38FHH71 pKa = 8.63KK72 pKa = 10.77EE73 pKa = 3.73NAIKK77 pKa = 10.41EE78 pKa = 4.28MAEE81 pKa = 3.79KK82 pKa = 10.36DD83 pKa = 3.81YY84 pKa = 11.22STVEE88 pKa = 3.87QNGG91 pKa = 3.19
Molecular weight: 10.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U6F5|A0A3Q8U6F5_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 35 OX=2494764 PE=4 SV=1
MM1 pKa = 7.7EE2 pKa = 5.3CLKK5 pKa = 10.1MIKK8 pKa = 8.7VTHH11 pKa = 6.58KK12 pKa = 10.62GVTHH16 pKa = 6.34TVPCGKK22 pKa = 10.08CAFCFSNKK30 pKa = 8.9RR31 pKa = 11.84SSWMFRR37 pKa = 11.84IHH39 pKa = 7.91HH40 pKa = 5.89EE41 pKa = 4.19MRR43 pKa = 11.84TQLHH47 pKa = 6.01KK48 pKa = 10.96GYY50 pKa = 9.96FLTLTYY56 pKa = 10.5DD57 pKa = 3.4EE58 pKa = 4.66KK59 pKa = 10.91HH60 pKa = 5.83VKK62 pKa = 8.59RR63 pKa = 11.84TAEE66 pKa = 3.83GRR68 pKa = 11.84LSLRR72 pKa = 11.84FRR74 pKa = 11.84DD75 pKa = 3.58VQLWLKK81 pKa = 9.12VLRR84 pKa = 11.84KK85 pKa = 9.02RR86 pKa = 11.84KK87 pKa = 10.14YY88 pKa = 7.37YY89 pKa = 9.82VKK91 pKa = 10.28YY92 pKa = 10.32IIVGEE97 pKa = 4.2YY98 pKa = 10.39GSEE101 pKa = 4.08TKK103 pKa = 10.34RR104 pKa = 11.84PHH106 pKa = 4.62YY107 pKa = 9.99HH108 pKa = 6.0ALIWTDD114 pKa = 3.73CAPEE118 pKa = 3.96MLEE121 pKa = 4.02KK122 pKa = 10.51LWFRR126 pKa = 11.84GRR128 pKa = 11.84IHH130 pKa = 7.2FGKK133 pKa = 8.77LTMASAMYY141 pKa = 7.83TLKK144 pKa = 11.14YY145 pKa = 10.1IIQPKK150 pKa = 9.76QKK152 pKa = 10.48DD153 pKa = 3.7EE154 pKa = 4.48NGVEE158 pKa = 4.02RR159 pKa = 11.84TRR161 pKa = 11.84AQFSKK166 pKa = 11.0GLGLAYY172 pKa = 10.56LDD174 pKa = 3.53TAAYY178 pKa = 9.96NYY180 pKa = 8.14HH181 pKa = 5.87TFDD184 pKa = 3.55YY185 pKa = 10.39DD186 pKa = 3.68SPEE189 pKa = 3.95MFSIIDD195 pKa = 3.76GKK197 pKa = 10.79KK198 pKa = 8.68VALPRR203 pKa = 11.84YY204 pKa = 8.04YY205 pKa = 10.0RR206 pKa = 11.84QKK208 pKa = 10.79IFTKK212 pKa = 10.52YY213 pKa = 10.11QLTKK217 pKa = 9.97HH218 pKa = 4.91AHH220 pKa = 5.44KK221 pKa = 10.72AKK223 pKa = 9.32WDD225 pKa = 3.98SIRR228 pKa = 11.84KK229 pKa = 8.22RR230 pKa = 11.84RR231 pKa = 11.84ARR233 pKa = 11.84MRR235 pKa = 11.84EE236 pKa = 3.72LKK238 pKa = 9.36MQGISNTKK246 pKa = 9.61VYY248 pKa = 9.42MHH250 pKa = 7.68ALRR253 pKa = 11.84VDD255 pKa = 3.17HH256 pKa = 7.26AKK258 pKa = 10.49RR259 pKa = 11.84IIEE262 pKa = 4.0NTKK265 pKa = 10.19YY266 pKa = 10.7GQSLL270 pKa = 3.48
MM1 pKa = 7.7EE2 pKa = 5.3CLKK5 pKa = 10.1MIKK8 pKa = 8.7VTHH11 pKa = 6.58KK12 pKa = 10.62GVTHH16 pKa = 6.34TVPCGKK22 pKa = 10.08CAFCFSNKK30 pKa = 8.9RR31 pKa = 11.84SSWMFRR37 pKa = 11.84IHH39 pKa = 7.91HH40 pKa = 5.89EE41 pKa = 4.19MRR43 pKa = 11.84TQLHH47 pKa = 6.01KK48 pKa = 10.96GYY50 pKa = 9.96FLTLTYY56 pKa = 10.5DD57 pKa = 3.4EE58 pKa = 4.66KK59 pKa = 10.91HH60 pKa = 5.83VKK62 pKa = 8.59RR63 pKa = 11.84TAEE66 pKa = 3.83GRR68 pKa = 11.84LSLRR72 pKa = 11.84FRR74 pKa = 11.84DD75 pKa = 3.58VQLWLKK81 pKa = 9.12VLRR84 pKa = 11.84KK85 pKa = 9.02RR86 pKa = 11.84KK87 pKa = 10.14YY88 pKa = 7.37YY89 pKa = 9.82VKK91 pKa = 10.28YY92 pKa = 10.32IIVGEE97 pKa = 4.2YY98 pKa = 10.39GSEE101 pKa = 4.08TKK103 pKa = 10.34RR104 pKa = 11.84PHH106 pKa = 4.62YY107 pKa = 9.99HH108 pKa = 6.0ALIWTDD114 pKa = 3.73CAPEE118 pKa = 3.96MLEE121 pKa = 4.02KK122 pKa = 10.51LWFRR126 pKa = 11.84GRR128 pKa = 11.84IHH130 pKa = 7.2FGKK133 pKa = 8.77LTMASAMYY141 pKa = 7.83TLKK144 pKa = 11.14YY145 pKa = 10.1IIQPKK150 pKa = 9.76QKK152 pKa = 10.48DD153 pKa = 3.7EE154 pKa = 4.48NGVEE158 pKa = 4.02RR159 pKa = 11.84TRR161 pKa = 11.84AQFSKK166 pKa = 11.0GLGLAYY172 pKa = 10.56LDD174 pKa = 3.53TAAYY178 pKa = 9.96NYY180 pKa = 8.14HH181 pKa = 5.87TFDD184 pKa = 3.55YY185 pKa = 10.39DD186 pKa = 3.68SPEE189 pKa = 3.95MFSIIDD195 pKa = 3.76GKK197 pKa = 10.79KK198 pKa = 8.68VALPRR203 pKa = 11.84YY204 pKa = 8.04YY205 pKa = 10.0RR206 pKa = 11.84QKK208 pKa = 10.79IFTKK212 pKa = 10.52YY213 pKa = 10.11QLTKK217 pKa = 9.97HH218 pKa = 4.91AHH220 pKa = 5.44KK221 pKa = 10.72AKK223 pKa = 9.32WDD225 pKa = 3.98SIRR228 pKa = 11.84KK229 pKa = 8.22RR230 pKa = 11.84RR231 pKa = 11.84ARR233 pKa = 11.84MRR235 pKa = 11.84EE236 pKa = 3.72LKK238 pKa = 9.36MQGISNTKK246 pKa = 9.61VYY248 pKa = 9.42MHH250 pKa = 7.68ALRR253 pKa = 11.84VDD255 pKa = 3.17HH256 pKa = 7.26AKK258 pKa = 10.49RR259 pKa = 11.84IIEE262 pKa = 4.0NTKK265 pKa = 10.19YY266 pKa = 10.7GQSLL270 pKa = 3.48
Molecular weight: 32.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1366 |
91 |
535 |
273.2 |
31.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.174 ± 0.505 | 0.586 ± 0.313 |
5.71 ± 0.796 | 6.442 ± 0.654 |
4.246 ± 0.642 | 6.735 ± 1.007 |
2.416 ± 0.655 | 5.198 ± 0.341 |
6.515 ± 1.826 | 7.687 ± 0.638 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.075 ± 0.337 | 4.319 ± 0.712 |
4.466 ± 0.941 | 4.905 ± 0.881 |
7.028 ± 0.73 | 5.71 ± 0.49 |
5.198 ± 0.462 | 6.296 ± 0.699 |
1.391 ± 0.276 | 4.905 ± 0.582 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
