
Flavobacterium rivuli WB 3.3-2 = DSM 21788
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; Flavobacterium rivuli
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
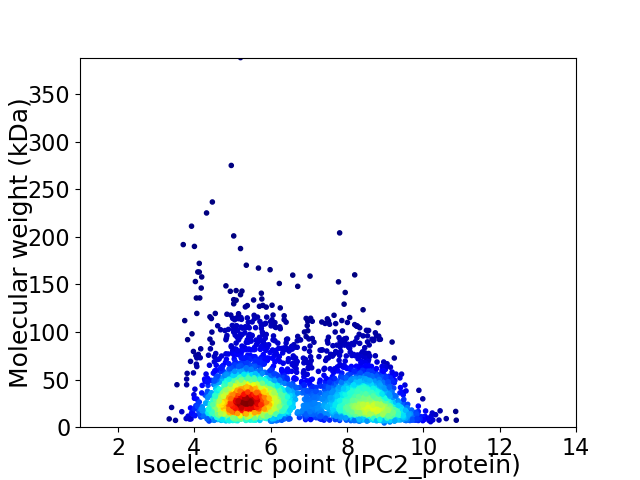
Virtual 2D-PAGE plot for 3691 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A2M1V0|A0A0A2M1V0_9FLAO Uncharacterized protein OS=Flavobacterium rivuli WB 3.3-2 = DSM 21788 OX=1121895 GN=Q765_11595 PE=4 SV=1
MM1 pKa = 7.39VLFAVLGGIVASCEE15 pKa = 4.2TEE17 pKa = 3.93PVDD20 pKa = 5.41PLLLTDD26 pKa = 5.25DD27 pKa = 4.3NGGTDD32 pKa = 4.16TIADD36 pKa = 3.75PGIFTVKK43 pKa = 10.1IGGTLFVADD52 pKa = 3.94STIATLGNGQIVIKK66 pKa = 10.2GYY68 pKa = 10.64KK69 pKa = 7.99GTEE72 pKa = 4.18GEE74 pKa = 4.31NVTITVQATTTGTYY88 pKa = 7.15TALMVYY94 pKa = 10.47DD95 pKa = 5.5PGTSDD100 pKa = 3.45NFYY103 pKa = 11.16SNISPASGPSGSATITTINTAARR126 pKa = 11.84VISGSFSFTGYY137 pKa = 11.0YY138 pKa = 10.19SDD140 pKa = 4.02TAQNLPNVAFTEE152 pKa = 5.11GIFTNISYY160 pKa = 11.03GGGTPPTNPSGPGVFTASVDD180 pKa = 3.78GAPFTAAVTNAVLGSGLFSIQGVDD204 pKa = 3.25ATGKK208 pKa = 9.91AISIAVEE215 pKa = 3.9EE216 pKa = 4.63TTPGTYY222 pKa = 9.35TGSQVLLAYY231 pKa = 10.12YY232 pKa = 10.3EE233 pKa = 4.22NATSEE238 pKa = 4.18DD239 pKa = 4.29SYY241 pKa = 12.1SNLLNDD247 pKa = 4.06TGTLTITSIDD257 pKa = 4.01TVNHH261 pKa = 6.9TITGTFSFTGDD272 pKa = 3.41YY273 pKa = 11.17SDD275 pKa = 4.12IDD277 pKa = 3.55NPRR280 pKa = 11.84TAKK283 pKa = 10.64VITSGVFTNVPYY295 pKa = 10.76VGSIDD300 pKa = 3.48NTDD303 pKa = 2.91IFTATVNGTAITYY316 pKa = 9.47GGSDD320 pKa = 3.7LVIGFIDD327 pKa = 3.63VNEE330 pKa = 4.65FSAVTLRR337 pKa = 11.84GTNATHH343 pKa = 7.25VIEE346 pKa = 4.23LQINEE351 pKa = 4.25DD352 pKa = 4.32TPVGTYY358 pKa = 10.32PFTDD362 pKa = 3.29ATNALPKK369 pKa = 10.0ATFTDD374 pKa = 4.25TSAPDD379 pKa = 4.73DD380 pKa = 3.21ITYY383 pKa = 10.76NITRR387 pKa = 11.84GSLVITANDD396 pKa = 3.69GVRR399 pKa = 11.84IKK401 pKa = 10.37GTFSFEE407 pKa = 3.76VRR409 pKa = 11.84NPANPNGAPLHH420 pKa = 5.63TVTGGTFDD428 pKa = 4.1IEE430 pKa = 4.32FF431 pKa = 4.31
MM1 pKa = 7.39VLFAVLGGIVASCEE15 pKa = 4.2TEE17 pKa = 3.93PVDD20 pKa = 5.41PLLLTDD26 pKa = 5.25DD27 pKa = 4.3NGGTDD32 pKa = 4.16TIADD36 pKa = 3.75PGIFTVKK43 pKa = 10.1IGGTLFVADD52 pKa = 3.94STIATLGNGQIVIKK66 pKa = 10.2GYY68 pKa = 10.64KK69 pKa = 7.99GTEE72 pKa = 4.18GEE74 pKa = 4.31NVTITVQATTTGTYY88 pKa = 7.15TALMVYY94 pKa = 10.47DD95 pKa = 5.5PGTSDD100 pKa = 3.45NFYY103 pKa = 11.16SNISPASGPSGSATITTINTAARR126 pKa = 11.84VISGSFSFTGYY137 pKa = 11.0YY138 pKa = 10.19SDD140 pKa = 4.02TAQNLPNVAFTEE152 pKa = 5.11GIFTNISYY160 pKa = 11.03GGGTPPTNPSGPGVFTASVDD180 pKa = 3.78GAPFTAAVTNAVLGSGLFSIQGVDD204 pKa = 3.25ATGKK208 pKa = 9.91AISIAVEE215 pKa = 3.9EE216 pKa = 4.63TTPGTYY222 pKa = 9.35TGSQVLLAYY231 pKa = 10.12YY232 pKa = 10.3EE233 pKa = 4.22NATSEE238 pKa = 4.18DD239 pKa = 4.29SYY241 pKa = 12.1SNLLNDD247 pKa = 4.06TGTLTITSIDD257 pKa = 4.01TVNHH261 pKa = 6.9TITGTFSFTGDD272 pKa = 3.41YY273 pKa = 11.17SDD275 pKa = 4.12IDD277 pKa = 3.55NPRR280 pKa = 11.84TAKK283 pKa = 10.64VITSGVFTNVPYY295 pKa = 10.76VGSIDD300 pKa = 3.48NTDD303 pKa = 2.91IFTATVNGTAITYY316 pKa = 9.47GGSDD320 pKa = 3.7LVIGFIDD327 pKa = 3.63VNEE330 pKa = 4.65FSAVTLRR337 pKa = 11.84GTNATHH343 pKa = 7.25VIEE346 pKa = 4.23LQINEE351 pKa = 4.25DD352 pKa = 4.32TPVGTYY358 pKa = 10.32PFTDD362 pKa = 3.29ATNALPKK369 pKa = 10.0ATFTDD374 pKa = 4.25TSAPDD379 pKa = 4.73DD380 pKa = 3.21ITYY383 pKa = 10.76NITRR387 pKa = 11.84GSLVITANDD396 pKa = 3.69GVRR399 pKa = 11.84IKK401 pKa = 10.37GTFSFEE407 pKa = 3.76VRR409 pKa = 11.84NPANPNGAPLHH420 pKa = 5.63TVTGGTFDD428 pKa = 4.1IEE430 pKa = 4.32FF431 pKa = 4.31
Molecular weight: 44.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A2M9B3|A0A0A2M9B3_9FLAO Uncharacterized protein OS=Flavobacterium rivuli WB 3.3-2 = DSM 21788 OX=1121895 GN=Q765_03940 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.39KK3 pKa = 10.22IIFTVAAIFAMGIASAQTNPTRR25 pKa = 11.84VPGTTTPTTTSPTTTSPTTTNGTINNGTNPNGTLNNGTTNGTLNNGVNNGTLNNPNTVPVPSTTTRR91 pKa = 11.84QNNNTRR97 pKa = 11.84STAIPGTTSPSTTSPSTTNNNAIRR121 pKa = 11.84PQTTASPSPTSTSTAVPSTSTRR143 pKa = 11.84MPASGTTTTTTTPSSTGTTPQPP165 pKa = 3.39
MM1 pKa = 7.46KK2 pKa = 10.39KK3 pKa = 10.22IIFTVAAIFAMGIASAQTNPTRR25 pKa = 11.84VPGTTTPTTTSPTTTSPTTTNGTINNGTNPNGTLNNGTTNGTLNNGVNNGTLNNPNTVPVPSTTTRR91 pKa = 11.84QNNNTRR97 pKa = 11.84STAIPGTTSPSTTSPSTTNNNAIRR121 pKa = 11.84PQTTASPSPTSTSTAVPSTSTRR143 pKa = 11.84MPASGTTTTTTTPSSTGTTPQPP165 pKa = 3.39
Molecular weight: 16.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1210709 |
41 |
3502 |
328.0 |
36.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.648 ± 0.043 | 0.753 ± 0.013 |
5.498 ± 0.03 | 5.912 ± 0.045 |
4.961 ± 0.032 | 6.743 ± 0.038 |
1.771 ± 0.019 | 7.401 ± 0.037 |
7.201 ± 0.05 | 9.011 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.018 | 5.952 ± 0.036 |
3.653 ± 0.025 | 3.441 ± 0.021 |
3.443 ± 0.025 | 5.982 ± 0.036 |
6.469 ± 0.064 | 6.568 ± 0.032 |
1.06 ± 0.017 | 4.299 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
