
Hypsizygus marmoreus (White beech mushroom) (Agaricus marmoreus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Lyophyllaceae; Hypsizygus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
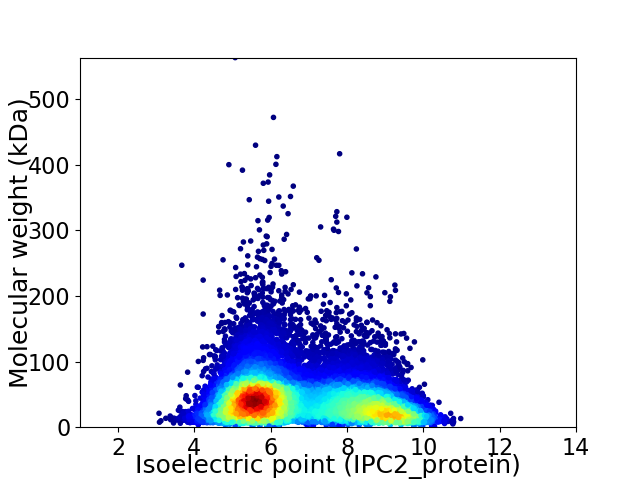
Virtual 2D-PAGE plot for 16572 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369JPG2|A0A369JPG2_HYPMA Uncharacterized protein OS=Hypsizygus marmoreus OX=39966 GN=Hypma_011511 PE=4 SV=1
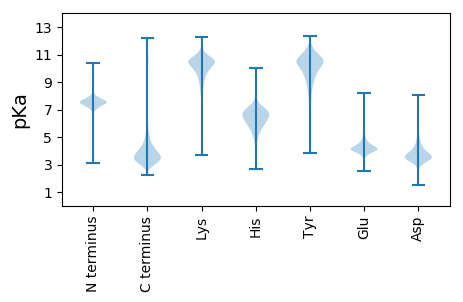
MM1 pKa = 7.26SANTISEE8 pKa = 4.5NGQHH12 pKa = 5.97TLPDD16 pKa = 3.85FQEE19 pKa = 4.1RR20 pKa = 11.84LATIRR25 pKa = 11.84TRR27 pKa = 11.84SRR29 pKa = 11.84ISYY32 pKa = 8.66TPVLTTPSLTTNTQQPQHH50 pKa = 7.36PDD52 pKa = 2.84TVMSDD57 pKa = 3.36TTNLFSLARR66 pKa = 11.84SKK68 pKa = 11.19LHH70 pKa = 5.81VSVGGKK76 pKa = 9.73DD77 pKa = 3.17SCSLHH82 pKa = 6.25RR83 pKa = 11.84WVLLKK88 pKa = 10.95NSIIRR93 pKa = 11.84SPSVPSSPSASDD105 pKa = 3.79YY106 pKa = 10.89PDD108 pKa = 3.38VNSEE112 pKa = 4.14YY113 pKa = 11.03AVDD116 pKa = 4.58DD117 pKa = 4.03EE118 pKa = 5.3ADD120 pKa = 3.58EE121 pKa = 4.41EE122 pKa = 4.76EE123 pKa = 4.46EE124 pKa = 4.48VNSEE128 pKa = 4.05EE129 pKa = 4.82LDD131 pKa = 3.41SFMFPDD137 pKa = 3.68AGKK140 pKa = 10.45LVGGSTADD148 pKa = 3.44VTASEE153 pKa = 4.96AEE155 pKa = 4.1WLDD158 pKa = 3.61SLLEE162 pKa = 4.04TLGDD166 pKa = 4.13DD167 pKa = 5.07DD168 pKa = 6.11DD169 pKa = 6.95DD170 pKa = 4.46EE171 pKa = 4.84FTVEE175 pKa = 4.49SDD177 pKa = 3.25PSLSILPVDD186 pKa = 4.4EE187 pKa = 6.87DD188 pKa = 4.87DD189 pKa = 5.48DD190 pKa = 4.23QLLLSPMGSPISSSDD205 pKa = 3.69DD206 pKa = 3.78LPPLSAFYY214 pKa = 9.9PHH216 pKa = 6.63STTVSYY222 pKa = 10.59SYY224 pKa = 9.69PYY226 pKa = 9.16PVPYY230 pKa = 10.25PPFHH234 pKa = 7.18PPLIHH239 pKa = 7.42SYY241 pKa = 10.88EE242 pKa = 4.07FDD244 pKa = 3.62SDD246 pKa = 4.03FDD248 pKa = 4.42PSLSSLPAPYY258 pKa = 10.12DD259 pKa = 3.64DD260 pKa = 4.91PLPYY264 pKa = 9.87HH265 pKa = 7.05DD266 pKa = 5.95LSDD269 pKa = 4.09SEE271 pKa = 4.4NLSVPDD277 pKa = 5.08AIEE280 pKa = 4.35DD281 pKa = 3.92TSDD284 pKa = 4.18DD285 pKa = 4.09EE286 pKa = 5.9SDD288 pKa = 4.0APPTPSMGRR297 pKa = 11.84STTSLTLLDD306 pKa = 4.34AASIPLPSEE315 pKa = 4.04RR316 pKa = 11.84SSLRR320 pKa = 11.84HH321 pKa = 5.44TNPHH325 pKa = 4.4VHH327 pKa = 5.86VEE329 pKa = 4.06SDD331 pKa = 3.13DD332 pKa = 4.07SYY334 pKa = 11.59FYY336 pKa = 10.6PFEE339 pKa = 5.38LDD341 pKa = 3.43PLPFPEE347 pKa = 5.35DD348 pKa = 2.75HH349 pKa = 6.48HH350 pKa = 7.05TYY352 pKa = 8.76NTYY355 pKa = 9.47QEE357 pKa = 4.49CC358 pKa = 3.69
MM1 pKa = 7.26SANTISEE8 pKa = 4.5NGQHH12 pKa = 5.97TLPDD16 pKa = 3.85FQEE19 pKa = 4.1RR20 pKa = 11.84LATIRR25 pKa = 11.84TRR27 pKa = 11.84SRR29 pKa = 11.84ISYY32 pKa = 8.66TPVLTTPSLTTNTQQPQHH50 pKa = 7.36PDD52 pKa = 2.84TVMSDD57 pKa = 3.36TTNLFSLARR66 pKa = 11.84SKK68 pKa = 11.19LHH70 pKa = 5.81VSVGGKK76 pKa = 9.73DD77 pKa = 3.17SCSLHH82 pKa = 6.25RR83 pKa = 11.84WVLLKK88 pKa = 10.95NSIIRR93 pKa = 11.84SPSVPSSPSASDD105 pKa = 3.79YY106 pKa = 10.89PDD108 pKa = 3.38VNSEE112 pKa = 4.14YY113 pKa = 11.03AVDD116 pKa = 4.58DD117 pKa = 4.03EE118 pKa = 5.3ADD120 pKa = 3.58EE121 pKa = 4.41EE122 pKa = 4.76EE123 pKa = 4.46EE124 pKa = 4.48VNSEE128 pKa = 4.05EE129 pKa = 4.82LDD131 pKa = 3.41SFMFPDD137 pKa = 3.68AGKK140 pKa = 10.45LVGGSTADD148 pKa = 3.44VTASEE153 pKa = 4.96AEE155 pKa = 4.1WLDD158 pKa = 3.61SLLEE162 pKa = 4.04TLGDD166 pKa = 4.13DD167 pKa = 5.07DD168 pKa = 6.11DD169 pKa = 6.95DD170 pKa = 4.46EE171 pKa = 4.84FTVEE175 pKa = 4.49SDD177 pKa = 3.25PSLSILPVDD186 pKa = 4.4EE187 pKa = 6.87DD188 pKa = 4.87DD189 pKa = 5.48DD190 pKa = 4.23QLLLSPMGSPISSSDD205 pKa = 3.69DD206 pKa = 3.78LPPLSAFYY214 pKa = 9.9PHH216 pKa = 6.63STTVSYY222 pKa = 10.59SYY224 pKa = 9.69PYY226 pKa = 9.16PVPYY230 pKa = 10.25PPFHH234 pKa = 7.18PPLIHH239 pKa = 7.42SYY241 pKa = 10.88EE242 pKa = 4.07FDD244 pKa = 3.62SDD246 pKa = 4.03FDD248 pKa = 4.42PSLSSLPAPYY258 pKa = 10.12DD259 pKa = 3.64DD260 pKa = 4.91PLPYY264 pKa = 9.87HH265 pKa = 7.05DD266 pKa = 5.95LSDD269 pKa = 4.09SEE271 pKa = 4.4NLSVPDD277 pKa = 5.08AIEE280 pKa = 4.35DD281 pKa = 3.92TSDD284 pKa = 4.18DD285 pKa = 4.09EE286 pKa = 5.9SDD288 pKa = 4.0APPTPSMGRR297 pKa = 11.84STTSLTLLDD306 pKa = 4.34AASIPLPSEE315 pKa = 4.04RR316 pKa = 11.84SSLRR320 pKa = 11.84HH321 pKa = 5.44TNPHH325 pKa = 4.4VHH327 pKa = 5.86VEE329 pKa = 4.06SDD331 pKa = 3.13DD332 pKa = 4.07SYY334 pKa = 11.59FYY336 pKa = 10.6PFEE339 pKa = 5.38LDD341 pKa = 3.43PLPFPEE347 pKa = 5.35DD348 pKa = 2.75HH349 pKa = 6.48HH350 pKa = 7.05TYY352 pKa = 8.76NTYY355 pKa = 9.47QEE357 pKa = 4.49CC358 pKa = 3.69
Molecular weight: 39.47 kDa
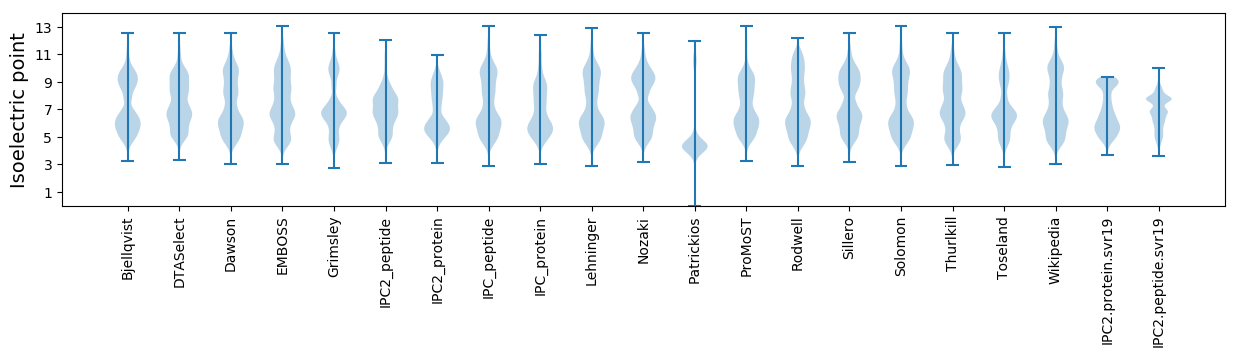
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369JI10|A0A369JI10_HYPMA Uncharacterized protein OS=Hypsizygus marmoreus OX=39966 GN=Hypma_012912 PE=4 SV=1
MM1 pKa = 7.61SSPTRR6 pKa = 11.84HH7 pKa = 5.77PTAASPPSLRR17 pKa = 11.84VTPLTPVTSLLRR29 pKa = 11.84AKK31 pKa = 10.23RR32 pKa = 11.84SAMLSAVPIRR42 pKa = 11.84PAPTTPAPAPTPSSTRR58 pKa = 11.84SLLASTAVLRR68 pKa = 11.84SRR70 pKa = 11.84ARR72 pKa = 11.84AAPWPSPDD80 pKa = 3.65KK81 pKa = 10.98TSLTAA86 pKa = 4.62
MM1 pKa = 7.61SSPTRR6 pKa = 11.84HH7 pKa = 5.77PTAASPPSLRR17 pKa = 11.84VTPLTPVTSLLRR29 pKa = 11.84AKK31 pKa = 10.23RR32 pKa = 11.84SAMLSAVPIRR42 pKa = 11.84PAPTTPAPAPTPSSTRR58 pKa = 11.84SLLASTAVLRR68 pKa = 11.84SRR70 pKa = 11.84ARR72 pKa = 11.84AAPWPSPDD80 pKa = 3.65KK81 pKa = 10.98TSLTAA86 pKa = 4.62
Molecular weight: 8.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7037948 |
8 |
5061 |
424.7 |
47.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.186 ± 0.017 | 1.292 ± 0.007 |
5.565 ± 0.012 | 5.711 ± 0.017 |
3.804 ± 0.01 | 6.273 ± 0.017 |
2.778 ± 0.01 | 4.995 ± 0.014 |
4.424 ± 0.016 | 9.335 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.062 ± 0.007 | 3.422 ± 0.01 |
6.884 ± 0.023 | 3.633 ± 0.012 |
6.342 ± 0.015 | 8.813 ± 0.024 |
6.219 ± 0.013 | 6.192 ± 0.013 |
1.434 ± 0.007 | 2.607 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
