
Octopus vulgaris (Common octopus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Cephalopoda; Coleoidea; Octopodiformes; Octopoda; Incirrata; Octopodidae; Octopus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
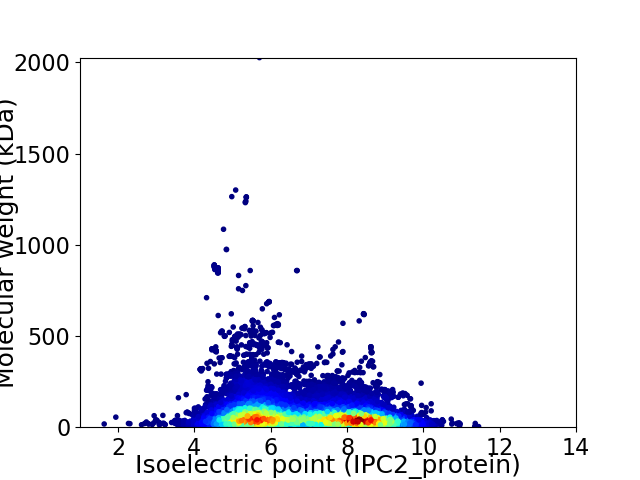
Virtual 2D-PAGE plot for 30330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A7E6F7M3|A0A7E6F7M3_OCTVU Isoform of A0A7E6F7L6 Non-specific serine/threonine protein kinase OS=Octopus vulgaris OX=6645 GN=LOC115217321 PE=3 SV=1
MM1 pKa = 7.39LQQIMSFLAIGEE13 pKa = 4.1AAKK16 pKa = 10.41YY17 pKa = 10.6VPGKK21 pKa = 9.75PKK23 pKa = 10.55LPPYY27 pKa = 10.3GQDD30 pKa = 3.25KK31 pKa = 11.13LLMAVPDD38 pKa = 4.08KK39 pKa = 10.57PCRR42 pKa = 11.84SSKK45 pKa = 10.57IEE47 pKa = 3.73WFLFTDD53 pKa = 4.28ADD55 pKa = 4.13NLGSWLEE62 pKa = 4.26AIKK65 pKa = 8.2TTLPQQPDD73 pKa = 3.51SSNGTPLCTEE83 pKa = 4.3TSVNMLPQPPFSTPPLQPLPPPPPPPGGPYY113 pKa = 10.23YY114 pKa = 10.78LPNTFGTNINRR125 pKa = 11.84APPSYY130 pKa = 10.32PPMYY134 pKa = 8.76QQQVTTISPVGYY146 pKa = 9.14PRR148 pKa = 11.84IQQPPPQTVIVRR160 pKa = 11.84EE161 pKa = 4.12EE162 pKa = 3.9RR163 pKa = 11.84SSNDD167 pKa = 2.41GFAPGLLLGDD177 pKa = 3.83ALGFGLGAGWFSGGGYY193 pKa = 10.2GGGYY197 pKa = 8.65TGFDD201 pKa = 3.66DD202 pKa = 4.88SQNIQINNYY211 pKa = 9.65YY212 pKa = 10.73DD213 pKa = 3.45VDD215 pKa = 3.62NTNIEE220 pKa = 4.23NTDD223 pKa = 3.8FTDD226 pKa = 2.99VDD228 pKa = 3.63NSFDD232 pKa = 3.89TNNFDD237 pKa = 4.6IGNNDD242 pKa = 3.45GFCDD246 pKa = 4.54DD247 pKa = 4.69GFNFF251 pKa = 4.53
MM1 pKa = 7.39LQQIMSFLAIGEE13 pKa = 4.1AAKK16 pKa = 10.41YY17 pKa = 10.6VPGKK21 pKa = 9.75PKK23 pKa = 10.55LPPYY27 pKa = 10.3GQDD30 pKa = 3.25KK31 pKa = 11.13LLMAVPDD38 pKa = 4.08KK39 pKa = 10.57PCRR42 pKa = 11.84SSKK45 pKa = 10.57IEE47 pKa = 3.73WFLFTDD53 pKa = 4.28ADD55 pKa = 4.13NLGSWLEE62 pKa = 4.26AIKK65 pKa = 8.2TTLPQQPDD73 pKa = 3.51SSNGTPLCTEE83 pKa = 4.3TSVNMLPQPPFSTPPLQPLPPPPPPPGGPYY113 pKa = 10.23YY114 pKa = 10.78LPNTFGTNINRR125 pKa = 11.84APPSYY130 pKa = 10.32PPMYY134 pKa = 8.76QQQVTTISPVGYY146 pKa = 9.14PRR148 pKa = 11.84IQQPPPQTVIVRR160 pKa = 11.84EE161 pKa = 4.12EE162 pKa = 3.9RR163 pKa = 11.84SSNDD167 pKa = 2.41GFAPGLLLGDD177 pKa = 3.83ALGFGLGAGWFSGGGYY193 pKa = 10.2GGGYY197 pKa = 8.65TGFDD201 pKa = 3.66DD202 pKa = 4.88SQNIQINNYY211 pKa = 9.65YY212 pKa = 10.73DD213 pKa = 3.45VDD215 pKa = 3.62NTNIEE220 pKa = 4.23NTDD223 pKa = 3.8FTDD226 pKa = 2.99VDD228 pKa = 3.63NSFDD232 pKa = 3.89TNNFDD237 pKa = 4.6IGNNDD242 pKa = 3.45GFCDD246 pKa = 4.54DD247 pKa = 4.69GFNFF251 pKa = 4.53
Molecular weight: 27.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P7SRX2|A0A6P7SRX2_OCTVU ubiquinol-cytochrome-c reductase complex assembly factor 1 isoform X1 OS=Octopus vulgaris OX=6645 GN=LOC115215843 PE=3 SV=1
MM1 pKa = 8.05PYY3 pKa = 10.02FADD6 pKa = 3.68EE7 pKa = 4.13VRR9 pKa = 11.84IKK11 pKa = 10.65QNSPIRR17 pKa = 11.84RR18 pKa = 11.84SSTTRR23 pKa = 11.84RR24 pKa = 11.84SSTTRR29 pKa = 11.84RR30 pKa = 11.84SSTILRR36 pKa = 11.84SSPTRR41 pKa = 11.84RR42 pKa = 11.84SSPTRR47 pKa = 11.84RR48 pKa = 11.84SSPTLRR54 pKa = 11.84SSPTRR59 pKa = 11.84RR60 pKa = 11.84SSPTRR65 pKa = 11.84RR66 pKa = 11.84SSTTRR71 pKa = 11.84RR72 pKa = 11.84SSLARR77 pKa = 11.84RR78 pKa = 11.84NSPIRR83 pKa = 11.84RR84 pKa = 11.84NSPIRR89 pKa = 11.84RR90 pKa = 11.84NSPTRR95 pKa = 11.84RR96 pKa = 11.84SSPTLRR102 pKa = 11.84NSPTLRR108 pKa = 11.84SSPIRR113 pKa = 11.84RR114 pKa = 11.84SSPIRR119 pKa = 11.84RR120 pKa = 11.84NSPIRR125 pKa = 11.84RR126 pKa = 11.84NSPTRR131 pKa = 11.84RR132 pKa = 11.84SSTTLRR138 pKa = 11.84NSPIRR143 pKa = 11.84RR144 pKa = 11.84SSPIRR149 pKa = 11.84RR150 pKa = 11.84SSPTRR155 pKa = 11.84RR156 pKa = 11.84SSPTRR161 pKa = 11.84RR162 pKa = 11.84SSPTRR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84SPTRR173 pKa = 11.84RR174 pKa = 11.84SSSFII179 pKa = 3.76
MM1 pKa = 8.05PYY3 pKa = 10.02FADD6 pKa = 3.68EE7 pKa = 4.13VRR9 pKa = 11.84IKK11 pKa = 10.65QNSPIRR17 pKa = 11.84RR18 pKa = 11.84SSTTRR23 pKa = 11.84RR24 pKa = 11.84SSTTRR29 pKa = 11.84RR30 pKa = 11.84SSTILRR36 pKa = 11.84SSPTRR41 pKa = 11.84RR42 pKa = 11.84SSPTRR47 pKa = 11.84RR48 pKa = 11.84SSPTLRR54 pKa = 11.84SSPTRR59 pKa = 11.84RR60 pKa = 11.84SSPTRR65 pKa = 11.84RR66 pKa = 11.84SSTTRR71 pKa = 11.84RR72 pKa = 11.84SSLARR77 pKa = 11.84RR78 pKa = 11.84NSPIRR83 pKa = 11.84RR84 pKa = 11.84NSPIRR89 pKa = 11.84RR90 pKa = 11.84NSPTRR95 pKa = 11.84RR96 pKa = 11.84SSPTLRR102 pKa = 11.84NSPTLRR108 pKa = 11.84SSPIRR113 pKa = 11.84RR114 pKa = 11.84SSPIRR119 pKa = 11.84RR120 pKa = 11.84NSPIRR125 pKa = 11.84RR126 pKa = 11.84NSPTRR131 pKa = 11.84RR132 pKa = 11.84SSTTLRR138 pKa = 11.84NSPIRR143 pKa = 11.84RR144 pKa = 11.84SSPIRR149 pKa = 11.84RR150 pKa = 11.84SSPTRR155 pKa = 11.84RR156 pKa = 11.84SSPTRR161 pKa = 11.84RR162 pKa = 11.84SSPTRR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84SPTRR173 pKa = 11.84RR174 pKa = 11.84SSSFII179 pKa = 3.76
Molecular weight: 20.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
20536984 |
9 |
18301 |
677.1 |
76.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.312 ± 0.013 | 2.245 ± 0.016 |
5.561 ± 0.016 | 6.734 ± 0.025 |
3.718 ± 0.011 | 5.148 ± 0.021 |
2.74 ± 0.013 | 5.555 ± 0.013 |
7.197 ± 0.018 | 8.47 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.199 ± 0.007 | 5.433 ± 0.017 |
5.05 ± 0.019 | 4.558 ± 0.016 |
4.637 ± 0.014 | 9.423 ± 0.028 |
6.351 ± 0.03 | 5.765 ± 0.012 |
0.908 ± 0.005 | 2.995 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
