
Capybara microvirus Cap1_SP_123
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
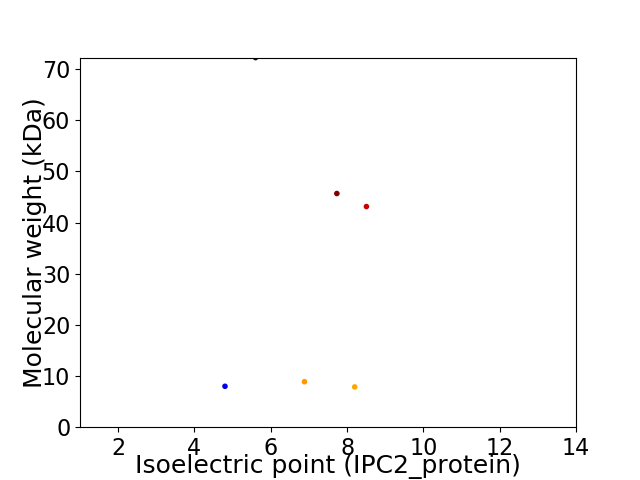
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
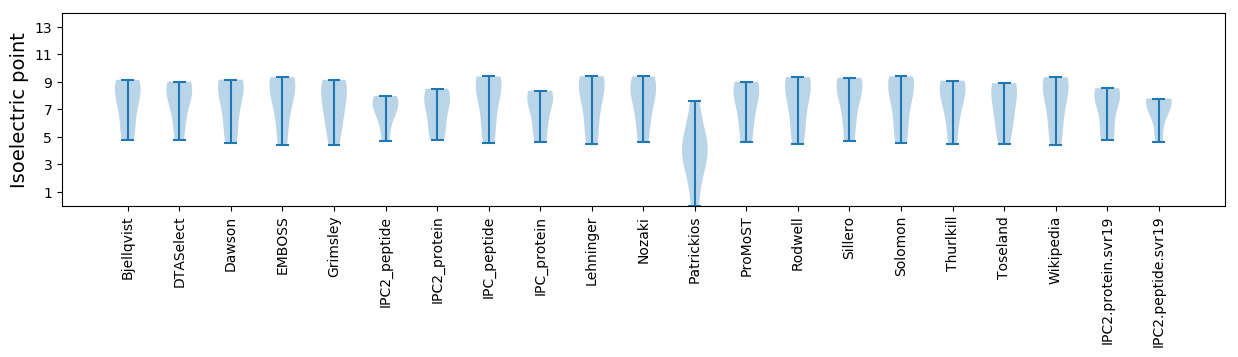
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVP2|A0A4V1FVP2_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_123 OX=2585378 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.45VFEE5 pKa = 4.64GFEE8 pKa = 4.47KK9 pKa = 10.55INSFRR14 pKa = 11.84IVAIPDD20 pKa = 3.35AKK22 pKa = 10.59RR23 pKa = 11.84LFEE26 pKa = 5.0LYY28 pKa = 9.68RR29 pKa = 11.84HH30 pKa = 5.97SPNLVGNEE38 pKa = 4.09EE39 pKa = 4.06EE40 pKa = 4.65TIDD43 pKa = 4.68LALEE47 pKa = 4.21SAQRR51 pKa = 11.84PTDD54 pKa = 3.37SMALYY59 pKa = 8.67EE60 pKa = 4.11QYY62 pKa = 11.0ARR64 pKa = 11.84SMDD67 pKa = 3.75SEE69 pKa = 4.41
MM1 pKa = 7.33KK2 pKa = 10.45VFEE5 pKa = 4.64GFEE8 pKa = 4.47KK9 pKa = 10.55INSFRR14 pKa = 11.84IVAIPDD20 pKa = 3.35AKK22 pKa = 10.59RR23 pKa = 11.84LFEE26 pKa = 5.0LYY28 pKa = 9.68RR29 pKa = 11.84HH30 pKa = 5.97SPNLVGNEE38 pKa = 4.09EE39 pKa = 4.06EE40 pKa = 4.65TIDD43 pKa = 4.68LALEE47 pKa = 4.21SAQRR51 pKa = 11.84PTDD54 pKa = 3.37SMALYY59 pKa = 8.67EE60 pKa = 4.11QYY62 pKa = 11.0ARR64 pKa = 11.84SMDD67 pKa = 3.75SEE69 pKa = 4.41
Molecular weight: 8.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W464|A0A4P8W464_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_123 OX=2585378 PE=4 SV=1
MM1 pKa = 7.71CISPRR6 pKa = 11.84IIPNPYY12 pKa = 10.33YY13 pKa = 10.34NDD15 pKa = 3.64KK16 pKa = 11.59GNTAWLHH23 pKa = 5.58DD24 pKa = 4.41TVCSHH29 pKa = 6.14IRR31 pKa = 11.84APCGHH36 pKa = 6.73CPEE39 pKa = 5.68CIHH42 pKa = 6.48NKK44 pKa = 8.01QNYY47 pKa = 6.13WVQRR51 pKa = 11.84CQMMEE56 pKa = 4.29FDD58 pKa = 4.91YY59 pKa = 10.81IPYY62 pKa = 9.35MVTLTYY68 pKa = 10.75NHH70 pKa = 7.31NIRR73 pKa = 11.84KK74 pKa = 8.74LHH76 pKa = 6.22IGDD79 pKa = 3.84YY80 pKa = 10.74SIMYY84 pKa = 9.9ADD86 pKa = 4.08KK87 pKa = 11.06TDD89 pKa = 3.23IQNMFKK95 pKa = 10.74RR96 pKa = 11.84MRR98 pKa = 11.84KK99 pKa = 7.78YY100 pKa = 10.82DD101 pKa = 3.34VFGRR105 pKa = 11.84EE106 pKa = 3.69FKK108 pKa = 10.98YY109 pKa = 10.39IAVSEE114 pKa = 4.33RR115 pKa = 11.84GKK117 pKa = 8.88EE118 pKa = 3.69THH120 pKa = 6.41RR121 pKa = 11.84PHH123 pKa = 5.09WHH125 pKa = 5.83ILFFLKK131 pKa = 10.52KK132 pKa = 9.92LANEE136 pKa = 4.37NEE138 pKa = 3.73WSYY141 pKa = 11.42ISHH144 pKa = 6.3EE145 pKa = 4.8KK146 pKa = 10.06EE147 pKa = 3.8LQSIFLNYY155 pKa = 6.24WTRR158 pKa = 11.84NYY160 pKa = 10.92GSTRR164 pKa = 11.84KK165 pKa = 9.24PDD167 pKa = 3.81YY168 pKa = 10.81KK169 pKa = 10.77PLLTLNKK176 pKa = 9.51RR177 pKa = 11.84RR178 pKa = 11.84TDD180 pKa = 2.92GRR182 pKa = 11.84RR183 pKa = 11.84NFDD186 pKa = 3.36LHH188 pKa = 7.77YY189 pKa = 10.59CMPGPSGNDD198 pKa = 2.86DD199 pKa = 3.4VLFYY203 pKa = 9.21CTKK206 pKa = 10.76YY207 pKa = 10.27ILKK210 pKa = 9.78HH211 pKa = 6.1DD212 pKa = 4.12PYY214 pKa = 10.97EE215 pKa = 4.7RR216 pKa = 11.84KK217 pKa = 9.48LHH219 pKa = 5.58SALMLNYY226 pKa = 9.99EE227 pKa = 3.89NDD229 pKa = 3.37EE230 pKa = 4.33AKK232 pKa = 10.75SYY234 pKa = 10.12WNTVKK239 pKa = 10.68SRR241 pKa = 11.84MLVSHH246 pKa = 6.69GFGTPYY252 pKa = 9.68TLSYY256 pKa = 10.52EE257 pKa = 4.13DD258 pKa = 4.31KK259 pKa = 10.59KK260 pKa = 11.36VKK262 pKa = 8.99KK263 pKa = 8.47TPNPSVIRR271 pKa = 11.84HH272 pKa = 4.97IRR274 pKa = 11.84KK275 pKa = 9.52GLDD278 pKa = 3.14LSTKK282 pKa = 8.62MPLYY286 pKa = 10.35INPNNGYY293 pKa = 7.86TMPLSPIYY301 pKa = 10.0KK302 pKa = 7.34PHH304 pKa = 6.57CMTRR308 pKa = 11.84EE309 pKa = 4.15DD310 pKa = 3.81YY311 pKa = 10.88QKK313 pKa = 10.78QFDD316 pKa = 4.33NNNLIDD322 pKa = 3.96INGDD326 pKa = 3.75KK327 pKa = 10.33INHH330 pKa = 6.25GSCFIDD336 pKa = 3.55TSTKK340 pKa = 10.22DD341 pKa = 3.23YY342 pKa = 11.3EE343 pKa = 4.61RR344 pKa = 11.84IIDD347 pKa = 3.88QFNHH351 pKa = 7.49RR352 pKa = 11.84ILQINKK358 pKa = 9.38HH359 pKa = 4.67YY360 pKa = 11.09
MM1 pKa = 7.71CISPRR6 pKa = 11.84IIPNPYY12 pKa = 10.33YY13 pKa = 10.34NDD15 pKa = 3.64KK16 pKa = 11.59GNTAWLHH23 pKa = 5.58DD24 pKa = 4.41TVCSHH29 pKa = 6.14IRR31 pKa = 11.84APCGHH36 pKa = 6.73CPEE39 pKa = 5.68CIHH42 pKa = 6.48NKK44 pKa = 8.01QNYY47 pKa = 6.13WVQRR51 pKa = 11.84CQMMEE56 pKa = 4.29FDD58 pKa = 4.91YY59 pKa = 10.81IPYY62 pKa = 9.35MVTLTYY68 pKa = 10.75NHH70 pKa = 7.31NIRR73 pKa = 11.84KK74 pKa = 8.74LHH76 pKa = 6.22IGDD79 pKa = 3.84YY80 pKa = 10.74SIMYY84 pKa = 9.9ADD86 pKa = 4.08KK87 pKa = 11.06TDD89 pKa = 3.23IQNMFKK95 pKa = 10.74RR96 pKa = 11.84MRR98 pKa = 11.84KK99 pKa = 7.78YY100 pKa = 10.82DD101 pKa = 3.34VFGRR105 pKa = 11.84EE106 pKa = 3.69FKK108 pKa = 10.98YY109 pKa = 10.39IAVSEE114 pKa = 4.33RR115 pKa = 11.84GKK117 pKa = 8.88EE118 pKa = 3.69THH120 pKa = 6.41RR121 pKa = 11.84PHH123 pKa = 5.09WHH125 pKa = 5.83ILFFLKK131 pKa = 10.52KK132 pKa = 9.92LANEE136 pKa = 4.37NEE138 pKa = 3.73WSYY141 pKa = 11.42ISHH144 pKa = 6.3EE145 pKa = 4.8KK146 pKa = 10.06EE147 pKa = 3.8LQSIFLNYY155 pKa = 6.24WTRR158 pKa = 11.84NYY160 pKa = 10.92GSTRR164 pKa = 11.84KK165 pKa = 9.24PDD167 pKa = 3.81YY168 pKa = 10.81KK169 pKa = 10.77PLLTLNKK176 pKa = 9.51RR177 pKa = 11.84RR178 pKa = 11.84TDD180 pKa = 2.92GRR182 pKa = 11.84RR183 pKa = 11.84NFDD186 pKa = 3.36LHH188 pKa = 7.77YY189 pKa = 10.59CMPGPSGNDD198 pKa = 2.86DD199 pKa = 3.4VLFYY203 pKa = 9.21CTKK206 pKa = 10.76YY207 pKa = 10.27ILKK210 pKa = 9.78HH211 pKa = 6.1DD212 pKa = 4.12PYY214 pKa = 10.97EE215 pKa = 4.7RR216 pKa = 11.84KK217 pKa = 9.48LHH219 pKa = 5.58SALMLNYY226 pKa = 9.99EE227 pKa = 3.89NDD229 pKa = 3.37EE230 pKa = 4.33AKK232 pKa = 10.75SYY234 pKa = 10.12WNTVKK239 pKa = 10.68SRR241 pKa = 11.84MLVSHH246 pKa = 6.69GFGTPYY252 pKa = 9.68TLSYY256 pKa = 10.52EE257 pKa = 4.13DD258 pKa = 4.31KK259 pKa = 10.59KK260 pKa = 11.36VKK262 pKa = 8.99KK263 pKa = 8.47TPNPSVIRR271 pKa = 11.84HH272 pKa = 4.97IRR274 pKa = 11.84KK275 pKa = 9.52GLDD278 pKa = 3.14LSTKK282 pKa = 8.62MPLYY286 pKa = 10.35INPNNGYY293 pKa = 7.86TMPLSPIYY301 pKa = 10.0KK302 pKa = 7.34PHH304 pKa = 6.57CMTRR308 pKa = 11.84EE309 pKa = 4.15DD310 pKa = 3.81YY311 pKa = 10.88QKK313 pKa = 10.78QFDD316 pKa = 4.33NNNLIDD322 pKa = 3.96INGDD326 pKa = 3.75KK327 pKa = 10.33INHH330 pKa = 6.25GSCFIDD336 pKa = 3.55TSTKK340 pKa = 10.22DD341 pKa = 3.23YY342 pKa = 11.3EE343 pKa = 4.61RR344 pKa = 11.84IIDD347 pKa = 3.88QFNHH351 pKa = 7.49RR352 pKa = 11.84ILQINKK358 pKa = 9.38HH359 pKa = 4.67YY360 pKa = 11.09
Molecular weight: 43.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1624 |
69 |
635 |
270.7 |
30.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.404 ± 1.905 | 1.478 ± 0.581 |
5.603 ± 0.577 | 5.172 ± 0.89 |
4.187 ± 1.124 | 4.372 ± 0.529 |
2.586 ± 0.948 | 6.897 ± 0.745 |
6.096 ± 1.452 | 7.451 ± 0.267 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.463 ± 0.36 | 8.067 ± 0.331 |
3.941 ± 0.596 | 4.31 ± 0.866 |
4.988 ± 0.526 | 8.682 ± 1.297 |
6.219 ± 1.2 | 4.31 ± 1.064 |
0.677 ± 0.296 | 6.096 ± 0.872 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
