
Ground squirrel hepatitis virus (strain 27) (GSHV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Orthohepadnavirus
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
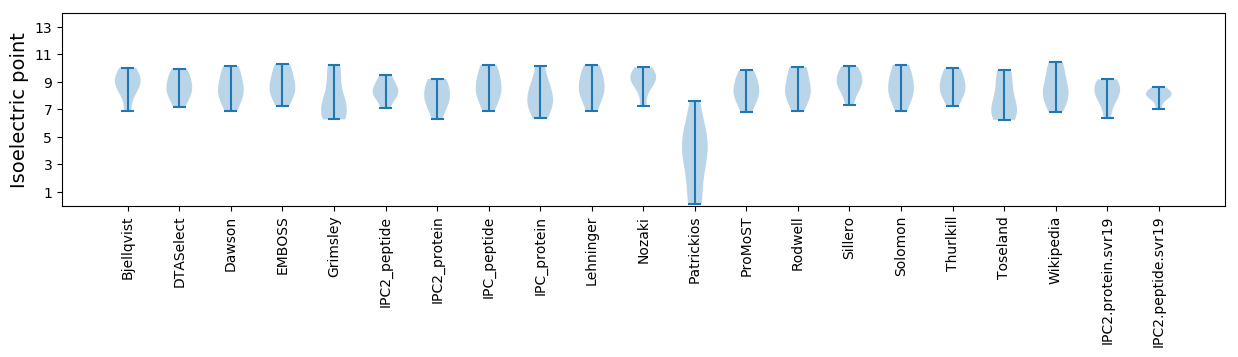
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03144-3|HBSAG-3_GSHV Isoform of P03144 Isoform S of Large envelope protein OS=Ground squirrel hepatitis virus (strain 27) OX=10406 GN=S PE=3 SV=1
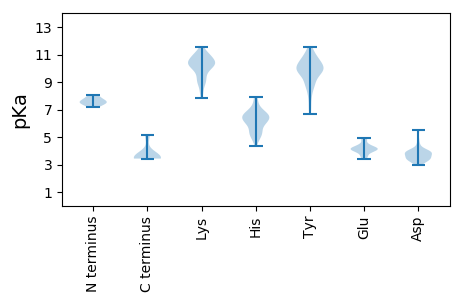
MM1 pKa = 7.6KK2 pKa = 10.24NQTGHH7 pKa = 6.09LQGFAEE13 pKa = 4.79GLRR16 pKa = 11.84ALTTSDD22 pKa = 3.51HH23 pKa = 6.63HH24 pKa = 6.52NSAYY28 pKa = 10.43GDD30 pKa = 3.82PFTTLSPVVPTVSTTLSPPLTIGDD54 pKa = 4.02PVLSTEE60 pKa = 4.3MSPSGLLGLLAGLQVVYY77 pKa = 8.81FLWTKK82 pKa = 10.34ILTIAQSLDD91 pKa = 3.07WWWTSLSFPGGIPEE105 pKa = 4.17CTGQNLQFQTCKK117 pKa = 10.28HH118 pKa = 6.29LPTSCPPTCNGFRR131 pKa = 11.84WMYY134 pKa = 9.79LRR136 pKa = 11.84RR137 pKa = 11.84FIIYY141 pKa = 10.35LLVLLLFLTFLLVLLDD157 pKa = 3.83WKK159 pKa = 11.02GLLPVCPMMPATEE172 pKa = 4.2TTVNCRR178 pKa = 11.84QCTISAQDD186 pKa = 3.43TFTTPYY192 pKa = 9.77CCCLKK197 pKa = 9.41PTAGNCTCWPIPSSWALGSYY217 pKa = 9.13LWEE220 pKa = 3.58WALARR225 pKa = 11.84FSWLSLLVPLLQWLGGISLTVWLLLIWMIWFWGPVLMSILPPFIPIFALFFLIWAYY281 pKa = 9.91II282 pKa = 3.43
MM1 pKa = 7.6KK2 pKa = 10.24NQTGHH7 pKa = 6.09LQGFAEE13 pKa = 4.79GLRR16 pKa = 11.84ALTTSDD22 pKa = 3.51HH23 pKa = 6.63HH24 pKa = 6.52NSAYY28 pKa = 10.43GDD30 pKa = 3.82PFTTLSPVVPTVSTTLSPPLTIGDD54 pKa = 4.02PVLSTEE60 pKa = 4.3MSPSGLLGLLAGLQVVYY77 pKa = 8.81FLWTKK82 pKa = 10.34ILTIAQSLDD91 pKa = 3.07WWWTSLSFPGGIPEE105 pKa = 4.17CTGQNLQFQTCKK117 pKa = 10.28HH118 pKa = 6.29LPTSCPPTCNGFRR131 pKa = 11.84WMYY134 pKa = 9.79LRR136 pKa = 11.84RR137 pKa = 11.84FIIYY141 pKa = 10.35LLVLLLFLTFLLVLLDD157 pKa = 3.83WKK159 pKa = 11.02GLLPVCPMMPATEE172 pKa = 4.2TTVNCRR178 pKa = 11.84QCTISAQDD186 pKa = 3.43TFTTPYY192 pKa = 9.77CCCLKK197 pKa = 9.41PTAGNCTCWPIPSSWALGSYY217 pKa = 9.13LWEE220 pKa = 3.58WALARR225 pKa = 11.84FSWLSLLVPLLQWLGGISLTVWLLLIWMIWFWGPVLMSILPPFIPIFALFFLIWAYY281 pKa = 9.91II282 pKa = 3.43
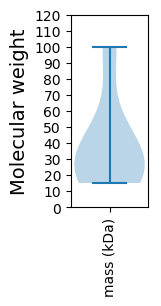
Molecular weight: 31.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03144-2|HBSAG-2_GSHV Isoform of P03144 Isoform M of Large envelope protein OS=Ground squirrel hepatitis virus (strain 27) OX=10406 GN=S PE=3 SV=1
MM1 pKa = 8.03DD2 pKa = 5.37IDD4 pKa = 4.05PYY6 pKa = 11.45KK7 pKa = 10.72EE8 pKa = 4.12FGSSYY13 pKa = 10.8QLLNFLPLDD22 pKa = 3.88FFPDD26 pKa = 3.91LNALVDD32 pKa = 3.96TAAALYY38 pKa = 10.25EE39 pKa = 4.12EE40 pKa = 4.9EE41 pKa = 4.22LTGRR45 pKa = 11.84EE46 pKa = 4.29HH47 pKa = 7.77CSPHH51 pKa = 5.3HH52 pKa = 4.74TAIRR56 pKa = 11.84QALVCWEE63 pKa = 4.11EE64 pKa = 4.01LTRR67 pKa = 11.84LITWMSEE74 pKa = 3.44NTTEE78 pKa = 3.99EE79 pKa = 3.73VRR81 pKa = 11.84RR82 pKa = 11.84IIVDD86 pKa = 3.38HH87 pKa = 6.85VNNTWGLKK95 pKa = 9.12VRR97 pKa = 11.84QTLWFHH103 pKa = 6.52LSCLTFGQHH112 pKa = 5.07TVQEE116 pKa = 4.43FLVSFGVWIRR126 pKa = 11.84TPAPYY131 pKa = 9.94RR132 pKa = 11.84PPNAPILSTLPEE144 pKa = 3.9HH145 pKa = 5.34TVIRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84GGSRR155 pKa = 11.84AARR158 pKa = 11.84SPRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84TPSPRR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84SQSPRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84SQSPASNCC187 pKa = 3.53
MM1 pKa = 8.03DD2 pKa = 5.37IDD4 pKa = 4.05PYY6 pKa = 11.45KK7 pKa = 10.72EE8 pKa = 4.12FGSSYY13 pKa = 10.8QLLNFLPLDD22 pKa = 3.88FFPDD26 pKa = 3.91LNALVDD32 pKa = 3.96TAAALYY38 pKa = 10.25EE39 pKa = 4.12EE40 pKa = 4.9EE41 pKa = 4.22LTGRR45 pKa = 11.84EE46 pKa = 4.29HH47 pKa = 7.77CSPHH51 pKa = 5.3HH52 pKa = 4.74TAIRR56 pKa = 11.84QALVCWEE63 pKa = 4.11EE64 pKa = 4.01LTRR67 pKa = 11.84LITWMSEE74 pKa = 3.44NTTEE78 pKa = 3.99EE79 pKa = 3.73VRR81 pKa = 11.84RR82 pKa = 11.84IIVDD86 pKa = 3.38HH87 pKa = 6.85VNNTWGLKK95 pKa = 9.12VRR97 pKa = 11.84QTLWFHH103 pKa = 6.52LSCLTFGQHH112 pKa = 5.07TVQEE116 pKa = 4.43FLVSFGVWIRR126 pKa = 11.84TPAPYY131 pKa = 9.94RR132 pKa = 11.84PPNAPILSTLPEE144 pKa = 3.9HH145 pKa = 5.34TVIRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84GGSRR155 pKa = 11.84AARR158 pKa = 11.84SPRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84TPSPRR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84SQSPRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84SQSPASNCC187 pKa = 3.53
Molecular weight: 21.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2355 |
138 |
881 |
336.4 |
38.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.605 ± 0.428 | 3.312 ± 0.443 |
3.057 ± 0.331 | 3.015 ± 0.558 |
5.011 ± 0.223 | 5.563 ± 0.393 |
2.718 ± 0.562 | 4.926 ± 0.429 |
3.142 ± 0.741 | 13.079 ± 1.146 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.741 ± 0.342 | 3.779 ± 0.714 |
7.771 ± 0.656 | 3.907 ± 0.102 |
5.902 ± 1.253 | 7.813 ± 0.547 |
7.983 ± 0.679 | 4.841 ± 0.179 |
3.864 ± 0.743 | 2.972 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
