
Eubacterium sp. CAG:76
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; environmental samples
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
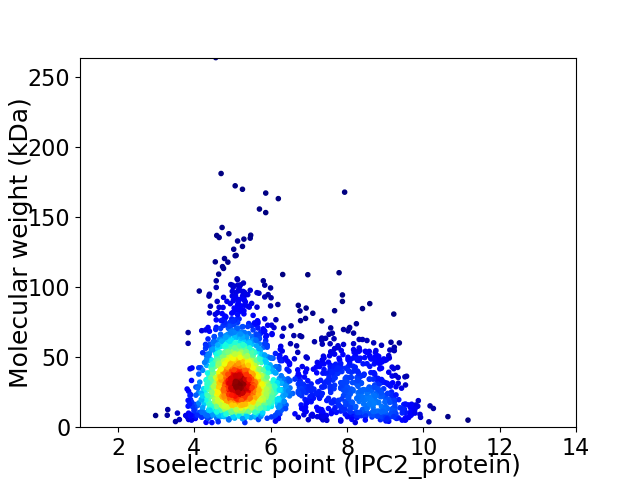
Virtual 2D-PAGE plot for 2278 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7NBV4|R7NBV4_9FIRM Putative manganese efflux pump MntP OS=Eubacterium sp. CAG:76 OX=1262892 GN=mntP PE=3 SV=1
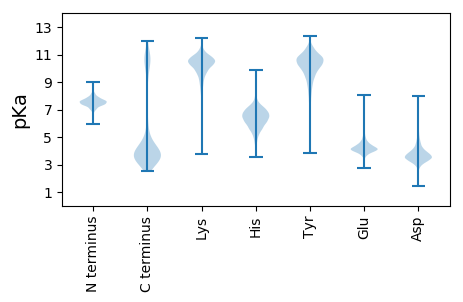
MM1 pKa = 7.38KK2 pKa = 10.26KK3 pKa = 10.33KK4 pKa = 10.32LVSLMLVAAMTASLTACGSKK24 pKa = 8.77KK25 pKa = 8.96TEE27 pKa = 4.04EE28 pKa = 4.3PTSTVHH34 pKa = 7.42KK35 pKa = 10.14SITAAEE41 pKa = 3.92YY42 pKa = 10.49DD43 pKa = 3.48AAITSDD49 pKa = 2.85AAIYY53 pKa = 10.13KK54 pKa = 10.12KK55 pKa = 10.53AFTMPEE61 pKa = 3.81YY62 pKa = 10.59KK63 pKa = 10.42GIQVTVDD70 pKa = 3.22KK71 pKa = 11.2SVLNVAEE78 pKa = 4.68SDD80 pKa = 3.3VDD82 pKa = 4.75DD83 pKa = 5.1YY84 pKa = 11.81INKK87 pKa = 10.0NIVSAEE93 pKa = 3.79ATTEE97 pKa = 4.01NVTSGVTADD106 pKa = 3.92GDD108 pKa = 4.54TVILDD113 pKa = 3.58YY114 pKa = 11.09SGKK117 pKa = 10.3LDD119 pKa = 3.68GEE121 pKa = 4.37AFSGGTATDD130 pKa = 3.3TTYY133 pKa = 10.8TIGSGNFISDD143 pKa = 3.68LDD145 pKa = 3.84KK146 pKa = 11.56GLVGLTVGQEE156 pKa = 3.74YY157 pKa = 10.23DD158 pKa = 3.64IPCTFPDD165 pKa = 4.27NYY167 pKa = 10.67TSDD170 pKa = 3.92LAGKK174 pKa = 9.59SVIFTVKK181 pKa = 8.57VTAIQKK187 pKa = 5.41TTYY190 pKa = 10.48PEE192 pKa = 4.03ITDD195 pKa = 3.11EE196 pKa = 3.95WVVNNKK202 pKa = 9.59EE203 pKa = 4.18SLNLSGDD210 pKa = 3.54TVADD214 pKa = 3.87FRR216 pKa = 11.84EE217 pKa = 4.24EE218 pKa = 3.56VRR220 pKa = 11.84KK221 pKa = 9.9IVEE224 pKa = 3.85ANANDD229 pKa = 3.89TYY231 pKa = 11.55LNNTFTSAYY240 pKa = 9.94QIISEE245 pKa = 4.46NIGDD249 pKa = 3.63VNYY252 pKa = 9.65PQDD255 pKa = 4.3EE256 pKa = 4.19IDD258 pKa = 4.1SLVEE262 pKa = 3.73VLNTNIEE269 pKa = 4.51SEE271 pKa = 4.31FNTYY275 pKa = 9.41GSYY278 pKa = 11.03YY279 pKa = 10.25GISDD283 pKa = 4.41LDD285 pKa = 3.85TYY287 pKa = 10.79KK288 pKa = 10.81KK289 pKa = 10.29SVYY292 pKa = 10.54GFDD295 pKa = 4.77SIDD298 pKa = 3.43AFNEE302 pKa = 4.1YY303 pKa = 9.08ATSSAQQYY311 pKa = 9.89LLQKK315 pKa = 10.1MIVTVIAADD324 pKa = 3.61NDD326 pKa = 3.46IHH328 pKa = 8.6VSEE331 pKa = 5.52DD332 pKa = 3.56EE333 pKa = 4.15INSYY337 pKa = 11.54GNDD340 pKa = 3.0LAQYY344 pKa = 10.15YY345 pKa = 10.76GYY347 pKa = 10.74DD348 pKa = 3.66DD349 pKa = 4.53FNAIVDD355 pKa = 3.97AVGSEE360 pKa = 4.32VVSEE364 pKa = 4.04IGYY367 pKa = 9.68EE368 pKa = 3.62ILYY371 pKa = 9.74QKK373 pKa = 10.5VVEE376 pKa = 4.92FEE378 pKa = 4.22CSQITEE384 pKa = 4.31VEE386 pKa = 4.1QQ387 pKa = 3.78
MM1 pKa = 7.38KK2 pKa = 10.26KK3 pKa = 10.33KK4 pKa = 10.32LVSLMLVAAMTASLTACGSKK24 pKa = 8.77KK25 pKa = 8.96TEE27 pKa = 4.04EE28 pKa = 4.3PTSTVHH34 pKa = 7.42KK35 pKa = 10.14SITAAEE41 pKa = 3.92YY42 pKa = 10.49DD43 pKa = 3.48AAITSDD49 pKa = 2.85AAIYY53 pKa = 10.13KK54 pKa = 10.12KK55 pKa = 10.53AFTMPEE61 pKa = 3.81YY62 pKa = 10.59KK63 pKa = 10.42GIQVTVDD70 pKa = 3.22KK71 pKa = 11.2SVLNVAEE78 pKa = 4.68SDD80 pKa = 3.3VDD82 pKa = 4.75DD83 pKa = 5.1YY84 pKa = 11.81INKK87 pKa = 10.0NIVSAEE93 pKa = 3.79ATTEE97 pKa = 4.01NVTSGVTADD106 pKa = 3.92GDD108 pKa = 4.54TVILDD113 pKa = 3.58YY114 pKa = 11.09SGKK117 pKa = 10.3LDD119 pKa = 3.68GEE121 pKa = 4.37AFSGGTATDD130 pKa = 3.3TTYY133 pKa = 10.8TIGSGNFISDD143 pKa = 3.68LDD145 pKa = 3.84KK146 pKa = 11.56GLVGLTVGQEE156 pKa = 3.74YY157 pKa = 10.23DD158 pKa = 3.64IPCTFPDD165 pKa = 4.27NYY167 pKa = 10.67TSDD170 pKa = 3.92LAGKK174 pKa = 9.59SVIFTVKK181 pKa = 8.57VTAIQKK187 pKa = 5.41TTYY190 pKa = 10.48PEE192 pKa = 4.03ITDD195 pKa = 3.11EE196 pKa = 3.95WVVNNKK202 pKa = 9.59EE203 pKa = 4.18SLNLSGDD210 pKa = 3.54TVADD214 pKa = 3.87FRR216 pKa = 11.84EE217 pKa = 4.24EE218 pKa = 3.56VRR220 pKa = 11.84KK221 pKa = 9.9IVEE224 pKa = 3.85ANANDD229 pKa = 3.89TYY231 pKa = 11.55LNNTFTSAYY240 pKa = 9.94QIISEE245 pKa = 4.46NIGDD249 pKa = 3.63VNYY252 pKa = 9.65PQDD255 pKa = 4.3EE256 pKa = 4.19IDD258 pKa = 4.1SLVEE262 pKa = 3.73VLNTNIEE269 pKa = 4.51SEE271 pKa = 4.31FNTYY275 pKa = 9.41GSYY278 pKa = 11.03YY279 pKa = 10.25GISDD283 pKa = 4.41LDD285 pKa = 3.85TYY287 pKa = 10.79KK288 pKa = 10.81KK289 pKa = 10.29SVYY292 pKa = 10.54GFDD295 pKa = 4.77SIDD298 pKa = 3.43AFNEE302 pKa = 4.1YY303 pKa = 9.08ATSSAQQYY311 pKa = 9.89LLQKK315 pKa = 10.1MIVTVIAADD324 pKa = 3.61NDD326 pKa = 3.46IHH328 pKa = 8.6VSEE331 pKa = 5.52DD332 pKa = 3.56EE333 pKa = 4.15INSYY337 pKa = 11.54GNDD340 pKa = 3.0LAQYY344 pKa = 10.15YY345 pKa = 10.76GYY347 pKa = 10.74DD348 pKa = 3.66DD349 pKa = 4.53FNAIVDD355 pKa = 3.97AVGSEE360 pKa = 4.32VVSEE364 pKa = 4.04IGYY367 pKa = 9.68EE368 pKa = 3.62ILYY371 pKa = 9.74QKK373 pKa = 10.5VVEE376 pKa = 4.92FEE378 pKa = 4.22CSQITEE384 pKa = 4.31VEE386 pKa = 4.1QQ387 pKa = 3.78
Molecular weight: 42.31 kDa
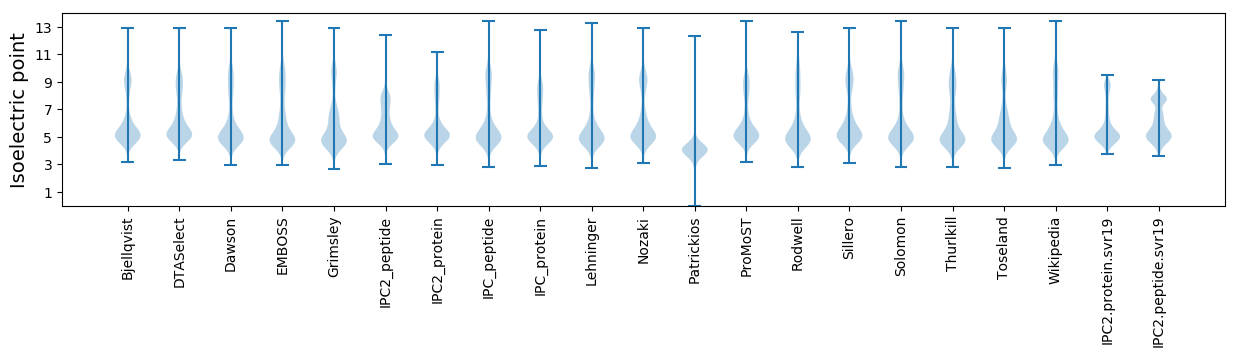
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7NBP4|R7NBP4_9FIRM tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase OS=Eubacterium sp. CAG:76 OX=1262892 GN=miaB PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.78TRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.82VHH16 pKa = 5.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.78TRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.82VHH16 pKa = 5.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
740048 |
29 |
2413 |
324.9 |
36.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.165 ± 0.049 | 1.488 ± 0.022 |
6.453 ± 0.041 | 6.786 ± 0.047 |
3.758 ± 0.038 | 6.596 ± 0.048 |
1.501 ± 0.022 | 8.579 ± 0.044 |
7.374 ± 0.045 | 8.048 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.108 ± 0.025 | 5.792 ± 0.043 |
2.729 ± 0.026 | 2.695 ± 0.026 |
3.798 ± 0.033 | 6.228 ± 0.049 |
5.426 ± 0.046 | 7.169 ± 0.046 |
0.713 ± 0.016 | 4.595 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
