
bank vole virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Narmovirus; Myodes narmovirus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
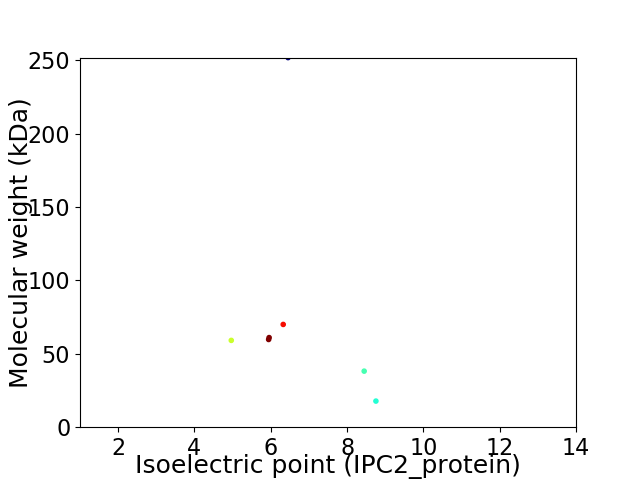
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4PJ44|A0A2H4PJ44_9MONO Attachment protein OS=bank vole virus 1 OX=2756244 GN=G PE=3 SV=1
MM1 pKa = 7.39SGVLSALKK9 pKa = 10.02EE10 pKa = 4.12FKK12 pKa = 10.03DD13 pKa = 3.25AKK15 pKa = 10.34LRR17 pKa = 11.84PKK19 pKa = 10.5HH20 pKa = 6.87DD21 pKa = 3.29GLTRR25 pKa = 11.84GAITSLKK32 pKa = 10.04HH33 pKa = 4.95KK34 pKa = 9.96VAVIVPGQEE43 pKa = 3.7GSKK46 pKa = 10.54VRR48 pKa = 11.84WQLLKK53 pKa = 11.14LLIGVAWSDD62 pKa = 3.2AANPSVQTGVMLSLISLISEE82 pKa = 4.21SPANMVRR89 pKa = 11.84RR90 pKa = 11.84LNNDD94 pKa = 3.08PDD96 pKa = 3.76LAITIVEE103 pKa = 3.91FTINSEE109 pKa = 4.08GEE111 pKa = 3.73LRR113 pKa = 11.84FASRR117 pKa = 11.84GMSYY121 pKa = 10.91DD122 pKa = 3.73DD123 pKa = 4.4QMSTYY128 pKa = 11.06LRR130 pKa = 11.84MRR132 pKa = 11.84DD133 pKa = 3.71NAPQAAQGEE142 pKa = 4.84HH143 pKa = 6.67PFEE146 pKa = 5.54EE147 pKa = 4.22SDD149 pKa = 4.03AWNQDD154 pKa = 3.88DD155 pKa = 5.66LPMDD159 pKa = 4.33EE160 pKa = 5.18YY161 pKa = 10.8MIANTTVQVQLWNLLIKK178 pKa = 10.59AVTAPDD184 pKa = 3.51TARR187 pKa = 11.84DD188 pKa = 3.61SEE190 pKa = 4.05QRR192 pKa = 11.84RR193 pKa = 11.84WLKK196 pKa = 9.9FVQQRR201 pKa = 11.84RR202 pKa = 11.84VEE204 pKa = 4.14AFYY207 pKa = 11.04KK208 pKa = 10.45LHH210 pKa = 6.82SIWADD215 pKa = 3.25KK216 pKa = 10.42VRR218 pKa = 11.84NLLAADD224 pKa = 4.28LSVRR228 pKa = 11.84RR229 pKa = 11.84YY230 pKa = 8.67MIRR233 pKa = 11.84TLIEE237 pKa = 3.86IQRR240 pKa = 11.84MGQTKK245 pKa = 9.91GRR247 pKa = 11.84LLEE250 pKa = 4.35VIADD254 pKa = 3.39IGNYY258 pKa = 9.27IEE260 pKa = 4.31EE261 pKa = 4.44SGLAGFQLTIRR272 pKa = 11.84YY273 pKa = 9.32GIEE276 pKa = 3.46TRR278 pKa = 11.84FAALALNEE286 pKa = 4.18FQGDD290 pKa = 3.56LATIEE295 pKa = 4.7RR296 pKa = 11.84LMKK299 pKa = 10.39LYY301 pKa = 11.16LEE303 pKa = 4.86MGPTAPFMVLLEE315 pKa = 5.44DD316 pKa = 3.55STQTKK321 pKa = 8.98FAPGNYY327 pKa = 8.46PLLWSYY333 pKa = 12.11AMGVGSALDD342 pKa = 3.12RR343 pKa = 11.84AMSNLNFNRR352 pKa = 11.84PYY354 pKa = 9.94MDD356 pKa = 2.96YY357 pKa = 11.0GYY359 pKa = 10.84FRR361 pKa = 11.84LGYY364 pKa = 9.84KK365 pKa = 9.74IVRR368 pKa = 11.84QSEE371 pKa = 4.17GSIDD375 pKa = 3.27SRR377 pKa = 11.84MANEE381 pKa = 5.13LEE383 pKa = 5.66LSPQDD388 pKa = 3.71QARR391 pKa = 11.84LRR393 pKa = 11.84ALVSGIGGRR402 pKa = 11.84EE403 pKa = 3.66DD404 pKa = 3.44VEE406 pKa = 4.49NIEE409 pKa = 4.12ARR411 pKa = 11.84GGNFQIADD419 pKa = 3.37IEE421 pKa = 4.51TTEE424 pKa = 4.08GEE426 pKa = 4.27EE427 pKa = 4.2LAGDD431 pKa = 4.03EE432 pKa = 4.22AQEE435 pKa = 4.05LPRR438 pKa = 11.84TRR440 pKa = 11.84RR441 pKa = 11.84TRR443 pKa = 11.84RR444 pKa = 11.84ARR446 pKa = 11.84SARR449 pKa = 11.84SSRR452 pKa = 11.84AQAGAGDD459 pKa = 5.02GININSYY466 pKa = 10.21SRR468 pKa = 11.84AVEE471 pKa = 3.72EE472 pKa = 4.24ALRR475 pKa = 11.84GSLTDD480 pKa = 5.25RR481 pKa = 11.84EE482 pKa = 4.46DD483 pKa = 3.3QSGDD487 pKa = 3.34QLYY490 pKa = 10.93NSPDD494 pKa = 3.3YY495 pKa = 9.79TRR497 pKa = 11.84EE498 pKa = 3.95MEE500 pKa = 4.23TEE502 pKa = 4.13EE503 pKa = 4.17EE504 pKa = 5.06DD505 pKa = 4.07EE506 pKa = 6.89DD507 pKa = 4.07EE508 pKa = 5.08GQDD511 pKa = 3.38NPLGLRR517 pKa = 11.84YY518 pKa = 9.7NDD520 pKa = 3.52EE521 pKa = 4.09SLLRR525 pKa = 4.7
MM1 pKa = 7.39SGVLSALKK9 pKa = 10.02EE10 pKa = 4.12FKK12 pKa = 10.03DD13 pKa = 3.25AKK15 pKa = 10.34LRR17 pKa = 11.84PKK19 pKa = 10.5HH20 pKa = 6.87DD21 pKa = 3.29GLTRR25 pKa = 11.84GAITSLKK32 pKa = 10.04HH33 pKa = 4.95KK34 pKa = 9.96VAVIVPGQEE43 pKa = 3.7GSKK46 pKa = 10.54VRR48 pKa = 11.84WQLLKK53 pKa = 11.14LLIGVAWSDD62 pKa = 3.2AANPSVQTGVMLSLISLISEE82 pKa = 4.21SPANMVRR89 pKa = 11.84RR90 pKa = 11.84LNNDD94 pKa = 3.08PDD96 pKa = 3.76LAITIVEE103 pKa = 3.91FTINSEE109 pKa = 4.08GEE111 pKa = 3.73LRR113 pKa = 11.84FASRR117 pKa = 11.84GMSYY121 pKa = 10.91DD122 pKa = 3.73DD123 pKa = 4.4QMSTYY128 pKa = 11.06LRR130 pKa = 11.84MRR132 pKa = 11.84DD133 pKa = 3.71NAPQAAQGEE142 pKa = 4.84HH143 pKa = 6.67PFEE146 pKa = 5.54EE147 pKa = 4.22SDD149 pKa = 4.03AWNQDD154 pKa = 3.88DD155 pKa = 5.66LPMDD159 pKa = 4.33EE160 pKa = 5.18YY161 pKa = 10.8MIANTTVQVQLWNLLIKK178 pKa = 10.59AVTAPDD184 pKa = 3.51TARR187 pKa = 11.84DD188 pKa = 3.61SEE190 pKa = 4.05QRR192 pKa = 11.84RR193 pKa = 11.84WLKK196 pKa = 9.9FVQQRR201 pKa = 11.84RR202 pKa = 11.84VEE204 pKa = 4.14AFYY207 pKa = 11.04KK208 pKa = 10.45LHH210 pKa = 6.82SIWADD215 pKa = 3.25KK216 pKa = 10.42VRR218 pKa = 11.84NLLAADD224 pKa = 4.28LSVRR228 pKa = 11.84RR229 pKa = 11.84YY230 pKa = 8.67MIRR233 pKa = 11.84TLIEE237 pKa = 3.86IQRR240 pKa = 11.84MGQTKK245 pKa = 9.91GRR247 pKa = 11.84LLEE250 pKa = 4.35VIADD254 pKa = 3.39IGNYY258 pKa = 9.27IEE260 pKa = 4.31EE261 pKa = 4.44SGLAGFQLTIRR272 pKa = 11.84YY273 pKa = 9.32GIEE276 pKa = 3.46TRR278 pKa = 11.84FAALALNEE286 pKa = 4.18FQGDD290 pKa = 3.56LATIEE295 pKa = 4.7RR296 pKa = 11.84LMKK299 pKa = 10.39LYY301 pKa = 11.16LEE303 pKa = 4.86MGPTAPFMVLLEE315 pKa = 5.44DD316 pKa = 3.55STQTKK321 pKa = 8.98FAPGNYY327 pKa = 8.46PLLWSYY333 pKa = 12.11AMGVGSALDD342 pKa = 3.12RR343 pKa = 11.84AMSNLNFNRR352 pKa = 11.84PYY354 pKa = 9.94MDD356 pKa = 2.96YY357 pKa = 11.0GYY359 pKa = 10.84FRR361 pKa = 11.84LGYY364 pKa = 9.84KK365 pKa = 9.74IVRR368 pKa = 11.84QSEE371 pKa = 4.17GSIDD375 pKa = 3.27SRR377 pKa = 11.84MANEE381 pKa = 5.13LEE383 pKa = 5.66LSPQDD388 pKa = 3.71QARR391 pKa = 11.84LRR393 pKa = 11.84ALVSGIGGRR402 pKa = 11.84EE403 pKa = 3.66DD404 pKa = 3.44VEE406 pKa = 4.49NIEE409 pKa = 4.12ARR411 pKa = 11.84GGNFQIADD419 pKa = 3.37IEE421 pKa = 4.51TTEE424 pKa = 4.08GEE426 pKa = 4.27EE427 pKa = 4.2LAGDD431 pKa = 4.03EE432 pKa = 4.22AQEE435 pKa = 4.05LPRR438 pKa = 11.84TRR440 pKa = 11.84RR441 pKa = 11.84TRR443 pKa = 11.84RR444 pKa = 11.84ARR446 pKa = 11.84SARR449 pKa = 11.84SSRR452 pKa = 11.84AQAGAGDD459 pKa = 5.02GININSYY466 pKa = 10.21SRR468 pKa = 11.84AVEE471 pKa = 3.72EE472 pKa = 4.24ALRR475 pKa = 11.84GSLTDD480 pKa = 5.25RR481 pKa = 11.84EE482 pKa = 4.46DD483 pKa = 3.3QSGDD487 pKa = 3.34QLYY490 pKa = 10.93NSPDD494 pKa = 3.3YY495 pKa = 9.79TRR497 pKa = 11.84EE498 pKa = 3.95MEE500 pKa = 4.23TEE502 pKa = 4.13EE503 pKa = 4.17EE504 pKa = 5.06DD505 pKa = 4.07EE506 pKa = 6.89DD507 pKa = 4.07EE508 pKa = 5.08GQDD511 pKa = 3.38NPLGLRR517 pKa = 11.84YY518 pKa = 9.7NDD520 pKa = 3.52EE521 pKa = 4.09SLLRR525 pKa = 4.7
Molecular weight: 59.13 kDa
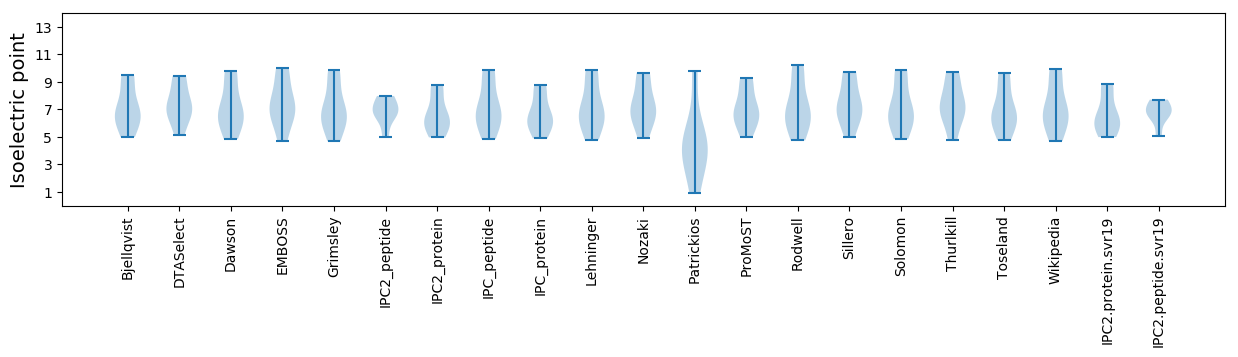
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4PJ60|A0A2H4PJ60_9MONO Isoform of A0A2H4PJ62 C protein OS=bank vole virus 1 OX=2756244 GN=P\V\C PE=4 SV=1
MM1 pKa = 7.8PSKK4 pKa = 10.31LWKK7 pKa = 9.79SLKK10 pKa = 9.81KK11 pKa = 10.58LRR13 pKa = 11.84VPRR16 pKa = 11.84PRR18 pKa = 11.84NNSVPEE24 pKa = 4.3STSPTPTSHH33 pKa = 7.58DD34 pKa = 3.21QTTDD38 pKa = 3.03PEE40 pKa = 4.17VRR42 pKa = 11.84VRR44 pKa = 11.84IGVRR48 pKa = 11.84RR49 pKa = 11.84NPDD52 pKa = 2.97LVGVEE57 pKa = 4.13RR58 pKa = 11.84QTKK61 pKa = 8.25VLKK64 pKa = 10.26QDD66 pKa = 3.5LALVILTQLRR76 pKa = 11.84DD77 pKa = 3.6LEE79 pKa = 4.53KK80 pKa = 10.4DD81 pKa = 3.6YY82 pKa = 10.59PAQVPHH88 pKa = 7.45IITSLKK94 pKa = 10.18YY95 pKa = 9.44STLQFVKK102 pKa = 9.48TILMRR107 pKa = 11.84VMEE110 pKa = 4.64GLPINSTWVRR120 pKa = 11.84QVEE123 pKa = 4.5EE124 pKa = 4.68YY125 pKa = 10.36ICTTSQEE132 pKa = 4.2VQNLKK137 pKa = 10.54EE138 pKa = 3.84AVEE141 pKa = 4.42WVRR144 pKa = 11.84KK145 pKa = 8.5MMEE148 pKa = 4.28DD149 pKa = 3.51QDD151 pKa = 3.88RR152 pKa = 11.84TT153 pKa = 3.67
MM1 pKa = 7.8PSKK4 pKa = 10.31LWKK7 pKa = 9.79SLKK10 pKa = 9.81KK11 pKa = 10.58LRR13 pKa = 11.84VPRR16 pKa = 11.84PRR18 pKa = 11.84NNSVPEE24 pKa = 4.3STSPTPTSHH33 pKa = 7.58DD34 pKa = 3.21QTTDD38 pKa = 3.03PEE40 pKa = 4.17VRR42 pKa = 11.84VRR44 pKa = 11.84IGVRR48 pKa = 11.84RR49 pKa = 11.84NPDD52 pKa = 2.97LVGVEE57 pKa = 4.13RR58 pKa = 11.84QTKK61 pKa = 8.25VLKK64 pKa = 10.26QDD66 pKa = 3.5LALVILTQLRR76 pKa = 11.84DD77 pKa = 3.6LEE79 pKa = 4.53KK80 pKa = 10.4DD81 pKa = 3.6YY82 pKa = 10.59PAQVPHH88 pKa = 7.45IITSLKK94 pKa = 10.18YY95 pKa = 9.44STLQFVKK102 pKa = 9.48TILMRR107 pKa = 11.84VMEE110 pKa = 4.64GLPINSTWVRR120 pKa = 11.84QVEE123 pKa = 4.5EE124 pKa = 4.68YY125 pKa = 10.36ICTTSQEE132 pKa = 4.2VQNLKK137 pKa = 10.54EE138 pKa = 3.84AVEE141 pKa = 4.42WVRR144 pKa = 11.84KK145 pKa = 8.5MMEE148 pKa = 4.28DD149 pKa = 3.51QDD151 pKa = 3.88RR152 pKa = 11.84TT153 pKa = 3.67
Molecular weight: 17.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4962 |
153 |
2206 |
708.9 |
79.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.784 ± 0.68 | 1.653 ± 0.276 |
5.361 ± 0.446 | 5.482 ± 0.537 |
3.083 ± 0.51 | 5.744 ± 0.582 |
2.559 ± 0.517 | 7.517 ± 0.691 |
5.038 ± 0.573 | 10.238 ± 0.559 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.257 ± 0.314 | 5.28 ± 0.442 |
4.938 ± 0.461 | 3.99 ± 0.423 |
5.401 ± 0.59 | 8.283 ± 0.716 |
6.308 ± 0.67 | 5.865 ± 0.772 |
1.028 ± 0.247 | 4.152 ± 0.653 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
