
Desulfuromonas soudanensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Desulfuromonadaceae; Desulfuromonas
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
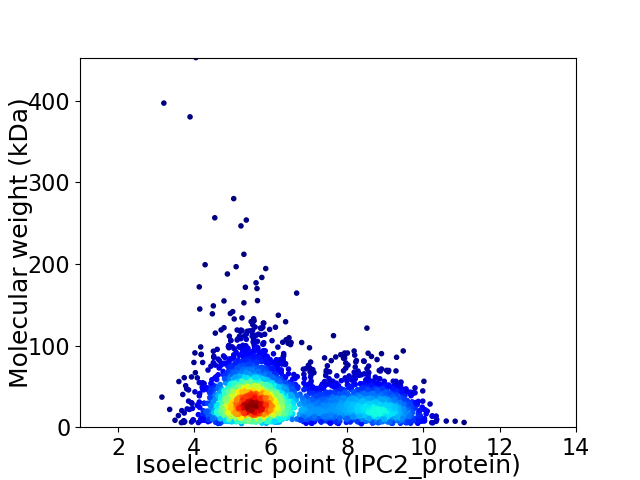
Virtual 2D-PAGE plot for 3469 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M3QFF4|A0A0M3QFF4_9DELT Autotransporter assembly factor TamA OS=Desulfuromonas soudanensis OX=1603606 GN=DSOUD_1192 PE=3 SV=1
MM1 pKa = 7.56SNLTPSPVIDD11 pKa = 4.72AGTDD15 pKa = 3.25QYY17 pKa = 12.07ALTGKK22 pKa = 9.5VVNLAGTSYY31 pKa = 11.32DD32 pKa = 3.6SAGGPLTYY40 pKa = 10.13QWNLVSTPSGSATTLSGANTLTPSFTADD68 pKa = 3.25LPGDD72 pKa = 3.83YY73 pKa = 10.49VLSLLADD80 pKa = 4.36DD81 pKa = 6.42GEE83 pKa = 4.65LQSQTDD89 pKa = 4.06TVTVHH94 pKa = 6.88ANVINCTFDD103 pKa = 3.38NPATLGMSGVIEE115 pKa = 4.18TSLSGDD121 pKa = 3.35IVPLCDD127 pKa = 3.43GWVLSANTTDD137 pKa = 5.03NRR139 pKa = 11.84VEE141 pKa = 3.98LQNVFTEE148 pKa = 4.12EE149 pKa = 4.19FEE151 pKa = 4.7RR152 pKa = 11.84SVSVDD157 pKa = 2.94GAVRR161 pKa = 11.84DD162 pKa = 3.84MEE164 pKa = 4.91FDD166 pKa = 3.99DD167 pKa = 5.75EE168 pKa = 5.21SGLLYY173 pKa = 10.42AVKK176 pKa = 10.28KK177 pKa = 9.93DD178 pKa = 3.57LPSISRR184 pKa = 11.84VDD186 pKa = 3.76MQAGTANLIPLPSLPLDD203 pKa = 3.8LALGGSNKK211 pKa = 9.91LFVSLRR217 pKa = 11.84SDD219 pKa = 3.5PLNIYY224 pKa = 10.55ADD226 pKa = 3.75YY227 pKa = 11.44SSISLVNGATGSVLKK242 pKa = 9.05TWPDD246 pKa = 3.18TSFSADD252 pKa = 3.97SILLAYY258 pKa = 9.92DD259 pKa = 3.63RR260 pKa = 11.84SGQQLVAGEE269 pKa = 4.2VWINPAEE276 pKa = 4.25LKK278 pKa = 10.41RR279 pKa = 11.84YY280 pKa = 10.09AFDD283 pKa = 4.06EE284 pKa = 4.78GALTLNLVDD293 pKa = 3.17QRR295 pKa = 11.84GYY297 pKa = 11.12VGTSAGDD304 pKa = 3.58LQISPDD310 pKa = 3.28GRR312 pKa = 11.84HH313 pKa = 5.71AAFTTGSGNGSVNPYY328 pKa = 9.79TIFDD332 pKa = 4.66FSTDD336 pKa = 3.31DD337 pKa = 3.28FGTVYY342 pKa = 11.09GEE344 pKa = 4.23WDD346 pKa = 3.14TDD348 pKa = 3.59AYY350 pKa = 9.42PLSGSFSPDD359 pKa = 2.17GNYY362 pKa = 9.37FVATNGSAIQLFDD375 pKa = 3.65VANHH379 pKa = 6.26NLLAEE384 pKa = 4.38YY385 pKa = 9.55PVSCNGLLEE394 pKa = 4.43TEE396 pKa = 4.18MVTRR400 pKa = 11.84FSRR403 pKa = 11.84GGNILFAYY411 pKa = 6.82GTCTGASSSTGRR423 pKa = 11.84IFWQVIEE430 pKa = 4.18PP431 pKa = 3.79
MM1 pKa = 7.56SNLTPSPVIDD11 pKa = 4.72AGTDD15 pKa = 3.25QYY17 pKa = 12.07ALTGKK22 pKa = 9.5VVNLAGTSYY31 pKa = 11.32DD32 pKa = 3.6SAGGPLTYY40 pKa = 10.13QWNLVSTPSGSATTLSGANTLTPSFTADD68 pKa = 3.25LPGDD72 pKa = 3.83YY73 pKa = 10.49VLSLLADD80 pKa = 4.36DD81 pKa = 6.42GEE83 pKa = 4.65LQSQTDD89 pKa = 4.06TVTVHH94 pKa = 6.88ANVINCTFDD103 pKa = 3.38NPATLGMSGVIEE115 pKa = 4.18TSLSGDD121 pKa = 3.35IVPLCDD127 pKa = 3.43GWVLSANTTDD137 pKa = 5.03NRR139 pKa = 11.84VEE141 pKa = 3.98LQNVFTEE148 pKa = 4.12EE149 pKa = 4.19FEE151 pKa = 4.7RR152 pKa = 11.84SVSVDD157 pKa = 2.94GAVRR161 pKa = 11.84DD162 pKa = 3.84MEE164 pKa = 4.91FDD166 pKa = 3.99DD167 pKa = 5.75EE168 pKa = 5.21SGLLYY173 pKa = 10.42AVKK176 pKa = 10.28KK177 pKa = 9.93DD178 pKa = 3.57LPSISRR184 pKa = 11.84VDD186 pKa = 3.76MQAGTANLIPLPSLPLDD203 pKa = 3.8LALGGSNKK211 pKa = 9.91LFVSLRR217 pKa = 11.84SDD219 pKa = 3.5PLNIYY224 pKa = 10.55ADD226 pKa = 3.75YY227 pKa = 11.44SSISLVNGATGSVLKK242 pKa = 9.05TWPDD246 pKa = 3.18TSFSADD252 pKa = 3.97SILLAYY258 pKa = 9.92DD259 pKa = 3.63RR260 pKa = 11.84SGQQLVAGEE269 pKa = 4.2VWINPAEE276 pKa = 4.25LKK278 pKa = 10.41RR279 pKa = 11.84YY280 pKa = 10.09AFDD283 pKa = 4.06EE284 pKa = 4.78GALTLNLVDD293 pKa = 3.17QRR295 pKa = 11.84GYY297 pKa = 11.12VGTSAGDD304 pKa = 3.58LQISPDD310 pKa = 3.28GRR312 pKa = 11.84HH313 pKa = 5.71AAFTTGSGNGSVNPYY328 pKa = 9.79TIFDD332 pKa = 4.66FSTDD336 pKa = 3.31DD337 pKa = 3.28FGTVYY342 pKa = 11.09GEE344 pKa = 4.23WDD346 pKa = 3.14TDD348 pKa = 3.59AYY350 pKa = 9.42PLSGSFSPDD359 pKa = 2.17GNYY362 pKa = 9.37FVATNGSAIQLFDD375 pKa = 3.65VANHH379 pKa = 6.26NLLAEE384 pKa = 4.38YY385 pKa = 9.55PVSCNGLLEE394 pKa = 4.43TEE396 pKa = 4.18MVTRR400 pKa = 11.84FSRR403 pKa = 11.84GGNILFAYY411 pKa = 6.82GTCTGASSSTGRR423 pKa = 11.84IFWQVIEE430 pKa = 4.18PP431 pKa = 3.79
Molecular weight: 45.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M3QER3|A0A0M3QER3_9DELT Chromosomal replication initiator protein DnaA OS=Desulfuromonas soudanensis OX=1603606 GN=dnaA PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.31QPSKK9 pKa = 9.13VKK11 pKa = 10.4RR12 pKa = 11.84KK13 pKa = 7.77RR14 pKa = 11.84THH16 pKa = 5.86GFRR19 pKa = 11.84KK20 pKa = 10.09RR21 pKa = 11.84MQTKK25 pKa = 9.99NGQSVLKK32 pKa = 10.26RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.49ILAATIPVKK49 pKa = 10.79
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.31QPSKK9 pKa = 9.13VKK11 pKa = 10.4RR12 pKa = 11.84KK13 pKa = 7.77RR14 pKa = 11.84THH16 pKa = 5.86GFRR19 pKa = 11.84KK20 pKa = 10.09RR21 pKa = 11.84MQTKK25 pKa = 9.99NGQSVLKK32 pKa = 10.26RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.49ILAATIPVKK49 pKa = 10.79
Molecular weight: 5.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1153148 |
40 |
4379 |
332.4 |
36.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.067 ± 0.059 | 1.163 ± 0.023 |
5.223 ± 0.037 | 6.761 ± 0.054 |
4.074 ± 0.031 | 8.659 ± 0.048 |
1.964 ± 0.023 | 5.426 ± 0.039 |
3.958 ± 0.047 | 11.457 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.138 ± 0.019 | 2.862 ± 0.045 |
4.9 ± 0.03 | 3.084 ± 0.025 |
6.815 ± 0.059 | 5.471 ± 0.033 |
5.156 ± 0.061 | 7.205 ± 0.036 |
1.042 ± 0.018 | 2.578 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
