
Cynomolgus cytomegalovirus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Cytomegalovirus; unclassified Cytomegalovirus
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
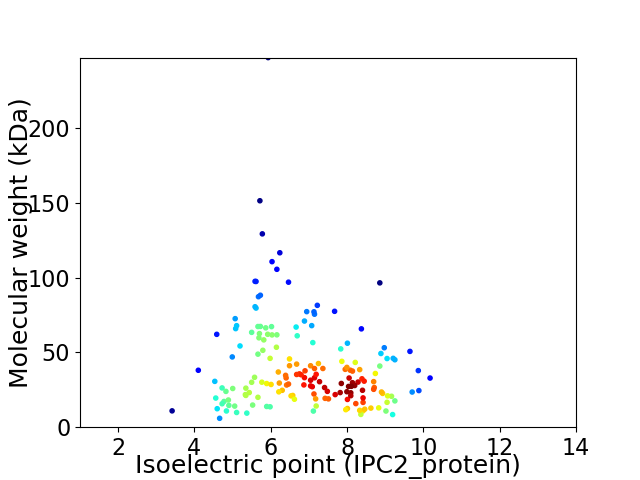
Virtual 2D-PAGE plot for 173 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6Z020|A0A1L6Z020_9BETA Capsid scaffolding protein OS=Cynomolgus cytomegalovirus OX=1919083 PE=3 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84WVAMRR7 pKa = 11.84WLLLIVATSAKK18 pKa = 10.29ASSTTAGNTSTPTASTITSTVTTASTTNMSTPSPASSPATTNTTTSSPSTKK69 pKa = 10.33SSTSSTNTTITTTGTASATTPKK91 pKa = 10.4SSTSVTSSPANSTSTSTKK109 pKa = 9.78PSTTNTSSPITTSSPASNTSTTTSSTSTVNGTNSTTEE146 pKa = 4.13TNTTSLPDD154 pKa = 3.27TTADD158 pKa = 3.45MNVTTTEE165 pKa = 4.5PYY167 pKa = 10.96NSTGYY172 pKa = 11.03EE173 pKa = 3.7NVTINITTPSPYY185 pKa = 10.0EE186 pKa = 3.61ALQIVDD192 pKa = 4.79LCNEE196 pKa = 4.43TISIVFKK203 pKa = 11.14DD204 pKa = 3.61PGEE207 pKa = 4.06EE208 pKa = 4.2DD209 pKa = 3.46EE210 pKa = 5.09SSEE213 pKa = 3.95TTEE216 pKa = 4.2YY217 pKa = 11.17SDD219 pKa = 4.21EE220 pKa = 4.3EE221 pKa = 4.3PVASSNEE228 pKa = 3.83DD229 pKa = 3.2DD230 pKa = 3.48STYY233 pKa = 10.3FPQSPGYY240 pKa = 7.88TLTYY244 pKa = 9.43DD245 pKa = 3.78TEE247 pKa = 4.17DD248 pKa = 3.27TIYY251 pKa = 10.65FQATCDD257 pKa = 4.08RR258 pKa = 11.84NDD260 pKa = 3.33TYY262 pKa = 11.56NITSCDD268 pKa = 3.68YY269 pKa = 10.21TNKK272 pKa = 10.2SVNSWSTVTSVSFFPPTLTPCHH294 pKa = 6.56KK295 pKa = 9.56PVAIIKK301 pKa = 9.88IGNDD305 pKa = 3.25SLVVSASATSNLVDD319 pKa = 5.76AIYY322 pKa = 10.99KK323 pKa = 10.49LLGLPDD329 pKa = 3.78VNSDD333 pKa = 4.72FINQLGRR340 pKa = 11.84YY341 pKa = 8.7HH342 pKa = 7.95PITLQGQIEE351 pKa = 4.19YY352 pKa = 9.97RR353 pKa = 11.84DD354 pKa = 3.61WYY356 pKa = 7.62TTEE359 pKa = 3.67
MM1 pKa = 7.5RR2 pKa = 11.84WVAMRR7 pKa = 11.84WLLLIVATSAKK18 pKa = 10.29ASSTTAGNTSTPTASTITSTVTTASTTNMSTPSPASSPATTNTTTSSPSTKK69 pKa = 10.33SSTSSTNTTITTTGTASATTPKK91 pKa = 10.4SSTSVTSSPANSTSTSTKK109 pKa = 9.78PSTTNTSSPITTSSPASNTSTTTSSTSTVNGTNSTTEE146 pKa = 4.13TNTTSLPDD154 pKa = 3.27TTADD158 pKa = 3.45MNVTTTEE165 pKa = 4.5PYY167 pKa = 10.96NSTGYY172 pKa = 11.03EE173 pKa = 3.7NVTINITTPSPYY185 pKa = 10.0EE186 pKa = 3.61ALQIVDD192 pKa = 4.79LCNEE196 pKa = 4.43TISIVFKK203 pKa = 11.14DD204 pKa = 3.61PGEE207 pKa = 4.06EE208 pKa = 4.2DD209 pKa = 3.46EE210 pKa = 5.09SSEE213 pKa = 3.95TTEE216 pKa = 4.2YY217 pKa = 11.17SDD219 pKa = 4.21EE220 pKa = 4.3EE221 pKa = 4.3PVASSNEE228 pKa = 3.83DD229 pKa = 3.2DD230 pKa = 3.48STYY233 pKa = 10.3FPQSPGYY240 pKa = 7.88TLTYY244 pKa = 9.43DD245 pKa = 3.78TEE247 pKa = 4.17DD248 pKa = 3.27TIYY251 pKa = 10.65FQATCDD257 pKa = 4.08RR258 pKa = 11.84NDD260 pKa = 3.33TYY262 pKa = 11.56NITSCDD268 pKa = 3.68YY269 pKa = 10.21TNKK272 pKa = 10.2SVNSWSTVTSVSFFPPTLTPCHH294 pKa = 6.56KK295 pKa = 9.56PVAIIKK301 pKa = 9.88IGNDD305 pKa = 3.25SLVVSASATSNLVDD319 pKa = 5.76AIYY322 pKa = 10.99KK323 pKa = 10.49LLGLPDD329 pKa = 3.78VNSDD333 pKa = 4.72FINQLGRR340 pKa = 11.84YY341 pKa = 8.7HH342 pKa = 7.95PITLQGQIEE351 pKa = 4.19YY352 pKa = 9.97RR353 pKa = 11.84DD354 pKa = 3.61WYY356 pKa = 7.62TTEE359 pKa = 3.67
Molecular weight: 38.05 kDa
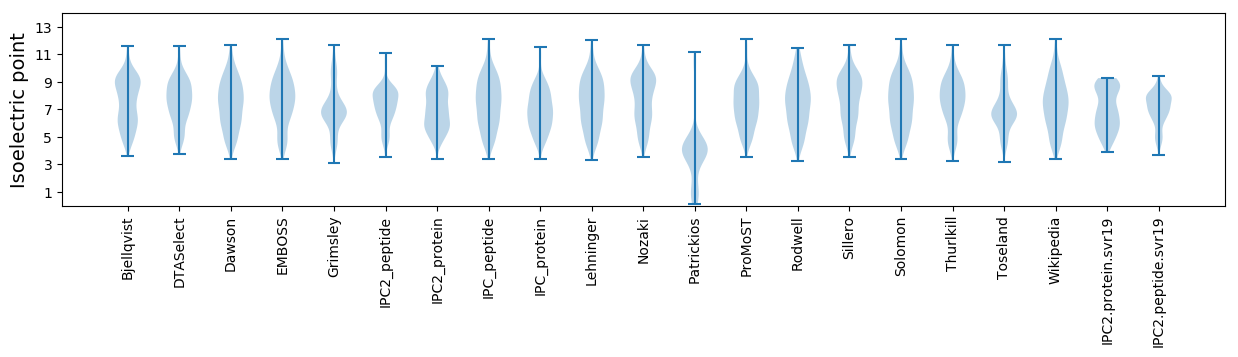
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6YZS4|A0A1L6YZS4_9BETA Cy50.1 OS=Cynomolgus cytomegalovirus OX=1919083 PE=4 SV=1
MM1 pKa = 7.56ARR3 pKa = 11.84HH4 pKa = 6.08RR5 pKa = 11.84CHH7 pKa = 6.77KK8 pKa = 10.42CWLLWLTVILGSWIHH23 pKa = 6.3LLNALEE29 pKa = 4.01IVEE32 pKa = 4.73EE33 pKa = 4.2YY34 pKa = 11.11VEE36 pKa = 4.07EE37 pKa = 4.24QEE39 pKa = 6.3IIMQTTRR46 pKa = 11.84QMALRR51 pKa = 11.84LAAAPVRR58 pKa = 11.84GRR60 pKa = 11.84RR61 pKa = 11.84QEE63 pKa = 3.52IDD65 pKa = 3.18GRR67 pKa = 11.84PPSIPSWLDD76 pKa = 2.93VRR78 pKa = 11.84SPRR81 pKa = 11.84PPRR84 pKa = 11.84RR85 pKa = 11.84HH86 pKa = 6.97DD87 pKa = 3.55EE88 pKa = 3.7NTMQRR93 pKa = 11.84NIRR96 pKa = 11.84WQHH99 pKa = 4.73EE100 pKa = 4.06QMQVLMSLHH109 pKa = 6.6HH110 pKa = 6.7SRR112 pKa = 11.84MQARR116 pKa = 11.84GWNPDD121 pKa = 2.48QRR123 pKa = 11.84TPDD126 pKa = 3.43RR127 pKa = 11.84LPAHH131 pKa = 7.21FGRR134 pKa = 11.84SASGSRR140 pKa = 11.84PVPIQRR146 pKa = 11.84IGTSTRR152 pKa = 11.84PTPMAEE158 pKa = 3.98EE159 pKa = 4.33AEE161 pKa = 4.45EE162 pKa = 3.91PRR164 pKa = 11.84VTIVRR169 pKa = 11.84NRR171 pKa = 11.84RR172 pKa = 11.84PDD174 pKa = 3.19NSSPGPRR181 pKa = 11.84LPHH184 pKa = 6.25KK185 pKa = 10.17RR186 pKa = 11.84RR187 pKa = 11.84NAVTFTGSTDD197 pKa = 3.48SLDD200 pKa = 3.57LTRR203 pKa = 11.84LRR205 pKa = 11.84LSYY208 pKa = 10.28EE209 pKa = 3.53RR210 pKa = 11.84RR211 pKa = 11.84HH212 pKa = 5.58RR213 pKa = 11.84KK214 pKa = 9.0GYY216 pKa = 9.28YY217 pKa = 8.18RR218 pKa = 11.84WKK220 pKa = 9.73HH221 pKa = 5.0HH222 pKa = 5.87GKK224 pKa = 10.33RR225 pKa = 11.84SPTVKK230 pKa = 10.49AHH232 pKa = 6.7IPPPRR237 pKa = 11.84PTISDD242 pKa = 3.26PMEE245 pKa = 4.18FRR247 pKa = 11.84PAPSPVPQRR256 pKa = 11.84RR257 pKa = 11.84TLNEE261 pKa = 3.77RR262 pKa = 11.84FNDD265 pKa = 3.38LLRR268 pKa = 11.84RR269 pKa = 11.84HH270 pKa = 6.03SAHH273 pKa = 6.35EE274 pKa = 4.2NIRR277 pKa = 11.84HH278 pKa = 5.58KK279 pKa = 10.27WHH281 pKa = 6.26VRR283 pKa = 11.84PKK285 pKa = 10.59DD286 pKa = 3.72DD287 pKa = 4.98PVWIYY292 pKa = 11.51NDD294 pKa = 3.88GEE296 pKa = 4.66SEE298 pKa = 4.18VTSIGEE304 pKa = 4.09RR305 pKa = 11.84ASLLTRR311 pKa = 11.84TRR313 pKa = 11.84SWIQSHH319 pKa = 5.41MHH321 pKa = 5.83RR322 pKa = 11.84TYY324 pKa = 9.33TWRR327 pKa = 11.84LGSSGGGNEE336 pKa = 3.81PLVRR340 pKa = 11.84SLSTEE345 pKa = 3.65QPRR348 pKa = 11.84NNNANQSRR356 pKa = 11.84LHH358 pKa = 6.33SPSEE362 pKa = 4.21SVQVNLEE369 pKa = 3.94TGEE372 pKa = 4.39VVVTCHH378 pKa = 4.92NTGANEE384 pKa = 3.98QQHH387 pKa = 6.01VLTSNQLCSRR397 pKa = 11.84NSMEE401 pKa = 4.59VTDD404 pKa = 3.94ACDD407 pKa = 3.14RR408 pKa = 11.84EE409 pKa = 4.66TPRR412 pKa = 11.84RR413 pKa = 11.84ALLGSSIALWLSGMWMRR430 pKa = 11.84HH431 pKa = 3.95YY432 pKa = 10.52RR433 pKa = 11.84KK434 pKa = 10.15SSS436 pKa = 2.93
MM1 pKa = 7.56ARR3 pKa = 11.84HH4 pKa = 6.08RR5 pKa = 11.84CHH7 pKa = 6.77KK8 pKa = 10.42CWLLWLTVILGSWIHH23 pKa = 6.3LLNALEE29 pKa = 4.01IVEE32 pKa = 4.73EE33 pKa = 4.2YY34 pKa = 11.11VEE36 pKa = 4.07EE37 pKa = 4.24QEE39 pKa = 6.3IIMQTTRR46 pKa = 11.84QMALRR51 pKa = 11.84LAAAPVRR58 pKa = 11.84GRR60 pKa = 11.84RR61 pKa = 11.84QEE63 pKa = 3.52IDD65 pKa = 3.18GRR67 pKa = 11.84PPSIPSWLDD76 pKa = 2.93VRR78 pKa = 11.84SPRR81 pKa = 11.84PPRR84 pKa = 11.84RR85 pKa = 11.84HH86 pKa = 6.97DD87 pKa = 3.55EE88 pKa = 3.7NTMQRR93 pKa = 11.84NIRR96 pKa = 11.84WQHH99 pKa = 4.73EE100 pKa = 4.06QMQVLMSLHH109 pKa = 6.6HH110 pKa = 6.7SRR112 pKa = 11.84MQARR116 pKa = 11.84GWNPDD121 pKa = 2.48QRR123 pKa = 11.84TPDD126 pKa = 3.43RR127 pKa = 11.84LPAHH131 pKa = 7.21FGRR134 pKa = 11.84SASGSRR140 pKa = 11.84PVPIQRR146 pKa = 11.84IGTSTRR152 pKa = 11.84PTPMAEE158 pKa = 3.98EE159 pKa = 4.33AEE161 pKa = 4.45EE162 pKa = 3.91PRR164 pKa = 11.84VTIVRR169 pKa = 11.84NRR171 pKa = 11.84RR172 pKa = 11.84PDD174 pKa = 3.19NSSPGPRR181 pKa = 11.84LPHH184 pKa = 6.25KK185 pKa = 10.17RR186 pKa = 11.84RR187 pKa = 11.84NAVTFTGSTDD197 pKa = 3.48SLDD200 pKa = 3.57LTRR203 pKa = 11.84LRR205 pKa = 11.84LSYY208 pKa = 10.28EE209 pKa = 3.53RR210 pKa = 11.84RR211 pKa = 11.84HH212 pKa = 5.58RR213 pKa = 11.84KK214 pKa = 9.0GYY216 pKa = 9.28YY217 pKa = 8.18RR218 pKa = 11.84WKK220 pKa = 9.73HH221 pKa = 5.0HH222 pKa = 5.87GKK224 pKa = 10.33RR225 pKa = 11.84SPTVKK230 pKa = 10.49AHH232 pKa = 6.7IPPPRR237 pKa = 11.84PTISDD242 pKa = 3.26PMEE245 pKa = 4.18FRR247 pKa = 11.84PAPSPVPQRR256 pKa = 11.84RR257 pKa = 11.84TLNEE261 pKa = 3.77RR262 pKa = 11.84FNDD265 pKa = 3.38LLRR268 pKa = 11.84RR269 pKa = 11.84HH270 pKa = 6.03SAHH273 pKa = 6.35EE274 pKa = 4.2NIRR277 pKa = 11.84HH278 pKa = 5.58KK279 pKa = 10.27WHH281 pKa = 6.26VRR283 pKa = 11.84PKK285 pKa = 10.59DD286 pKa = 3.72DD287 pKa = 4.98PVWIYY292 pKa = 11.51NDD294 pKa = 3.88GEE296 pKa = 4.66SEE298 pKa = 4.18VTSIGEE304 pKa = 4.09RR305 pKa = 11.84ASLLTRR311 pKa = 11.84TRR313 pKa = 11.84SWIQSHH319 pKa = 5.41MHH321 pKa = 5.83RR322 pKa = 11.84TYY324 pKa = 9.33TWRR327 pKa = 11.84LGSSGGGNEE336 pKa = 3.81PLVRR340 pKa = 11.84SLSTEE345 pKa = 3.65QPRR348 pKa = 11.84NNNANQSRR356 pKa = 11.84LHH358 pKa = 6.33SPSEE362 pKa = 4.21SVQVNLEE369 pKa = 3.94TGEE372 pKa = 4.39VVVTCHH378 pKa = 4.92NTGANEE384 pKa = 3.98QQHH387 pKa = 6.01VLTSNQLCSRR397 pKa = 11.84NSMEE401 pKa = 4.59VTDD404 pKa = 3.94ACDD407 pKa = 3.14RR408 pKa = 11.84EE409 pKa = 4.66TPRR412 pKa = 11.84RR413 pKa = 11.84ALLGSSIALWLSGMWMRR430 pKa = 11.84HH431 pKa = 3.95YY432 pKa = 10.52RR433 pKa = 11.84KK434 pKa = 10.15SSS436 pKa = 2.93
Molecular weight: 50.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
62217 |
53 |
2179 |
359.6 |
40.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.582 ± 0.144 | 2.676 ± 0.117 |
4.79 ± 0.139 | 5.235 ± 0.158 |
4.121 ± 0.117 | 4.568 ± 0.125 |
3.025 ± 0.116 | 5.174 ± 0.125 |
3.874 ± 0.143 | 9.88 ± 0.198 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.589 ± 0.089 | 4.285 ± 0.108 |
5.526 ± 0.204 | 3.893 ± 0.157 |
6.379 ± 0.153 | 7.948 ± 0.175 |
7.389 ± 0.271 | 6.961 ± 0.163 |
1.385 ± 0.077 | 3.721 ± 0.092 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
