
Thrips palmi (Melon thrips)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Paraneoptera; Thysanoptera; Terebrantia; Thripoidea; Thripidae; Thripinae; Thrips
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
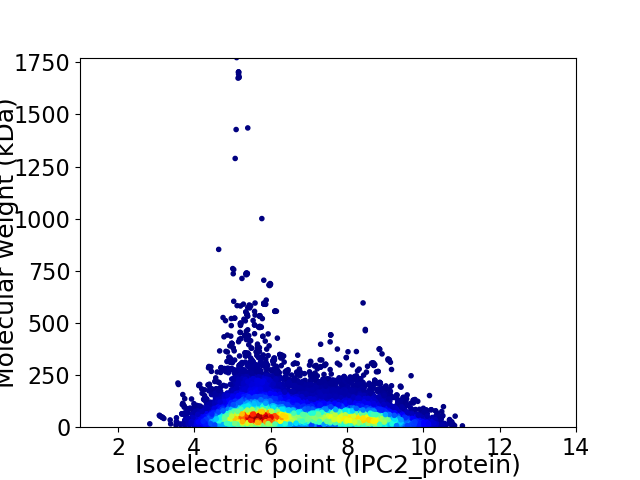
Virtual 2D-PAGE plot for 20971 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P8ZIQ7|A0A6P8ZIQ7_THRPL Isoform of A0A6P8YJ99 Regulatory protein zeste OS=Thrips palmi OX=161013 GN=LOC117641037 PE=4 SV=1
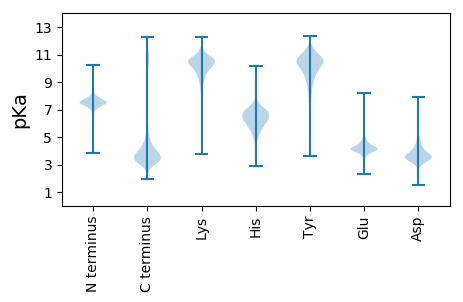
MM1 pKa = 7.12EE2 pKa = 4.83TLDD5 pKa = 3.6TALSFSVFPPTPPPSTSSPSASSGSLWDD33 pKa = 3.57ADD35 pKa = 3.35LWGASSATWLDD46 pKa = 3.44EE47 pKa = 3.89GWALDD52 pKa = 3.7GLPDD56 pKa = 3.71VKK58 pKa = 10.74PDD60 pKa = 3.63VSEE63 pKa = 4.43LLQASLRR70 pKa = 11.84QDD72 pKa = 3.91LPVLNADD79 pKa = 3.48STTFLLDD86 pKa = 3.2GDD88 pKa = 4.51AAPAPAAWLPQWSYY102 pKa = 11.37CAYY105 pKa = 8.85CAAGGGVGAAGCGASVSQPLPPQGPSQLYY134 pKa = 8.64PDD136 pKa = 3.75QGADD140 pKa = 3.09QGADD144 pKa = 3.03QGGGYY149 pKa = 10.46LVDD152 pKa = 4.95PYY154 pKa = 10.96LGTLAGPMPAPFADD168 pKa = 3.32GLVFVDD174 pKa = 5.83SPALCPVLSTVPEE187 pKa = 4.27DD188 pKa = 3.48TCLEE192 pKa = 4.43DD193 pKa = 3.5AVHH196 pKa = 6.81WANPVPPAPSPLPAEE211 pKa = 4.31LSGHH215 pKa = 6.53RR216 pKa = 11.84SALPLPEE223 pKa = 4.89PPGKK227 pKa = 9.86SSKK230 pKa = 10.02RR231 pKa = 11.84APRR234 pKa = 11.84TSRR237 pKa = 11.84DD238 pKa = 3.26PFII241 pKa = 6.05
MM1 pKa = 7.12EE2 pKa = 4.83TLDD5 pKa = 3.6TALSFSVFPPTPPPSTSSPSASSGSLWDD33 pKa = 3.57ADD35 pKa = 3.35LWGASSATWLDD46 pKa = 3.44EE47 pKa = 3.89GWALDD52 pKa = 3.7GLPDD56 pKa = 3.71VKK58 pKa = 10.74PDD60 pKa = 3.63VSEE63 pKa = 4.43LLQASLRR70 pKa = 11.84QDD72 pKa = 3.91LPVLNADD79 pKa = 3.48STTFLLDD86 pKa = 3.2GDD88 pKa = 4.51AAPAPAAWLPQWSYY102 pKa = 11.37CAYY105 pKa = 8.85CAAGGGVGAAGCGASVSQPLPPQGPSQLYY134 pKa = 8.64PDD136 pKa = 3.75QGADD140 pKa = 3.09QGADD144 pKa = 3.03QGGGYY149 pKa = 10.46LVDD152 pKa = 4.95PYY154 pKa = 10.96LGTLAGPMPAPFADD168 pKa = 3.32GLVFVDD174 pKa = 5.83SPALCPVLSTVPEE187 pKa = 4.27DD188 pKa = 3.48TCLEE192 pKa = 4.43DD193 pKa = 3.5AVHH196 pKa = 6.81WANPVPPAPSPLPAEE211 pKa = 4.31LSGHH215 pKa = 6.53RR216 pKa = 11.84SALPLPEE223 pKa = 4.89PPGKK227 pKa = 9.86SSKK230 pKa = 10.02RR231 pKa = 11.84APRR234 pKa = 11.84TSRR237 pKa = 11.84DD238 pKa = 3.26PFII241 pKa = 6.05
Molecular weight: 24.73 kDa
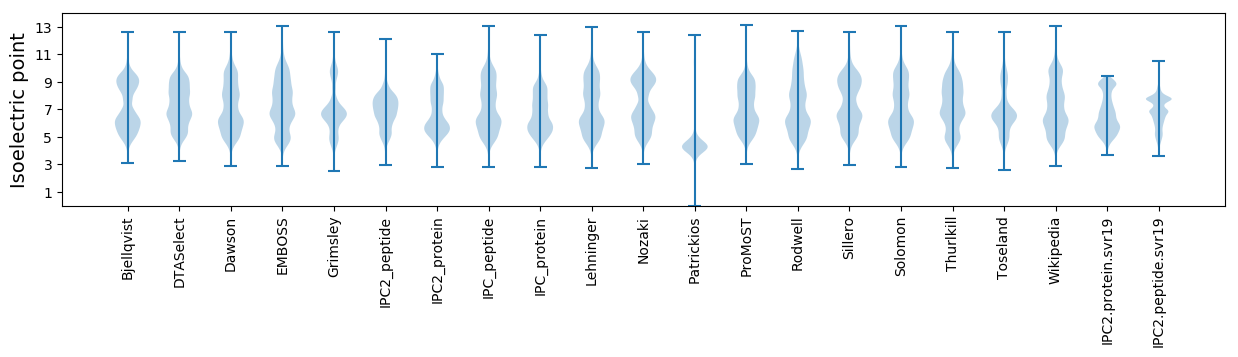
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P9AEV0|A0A6P9AEV0_THRPL TOX high mobility group box family member 4-like isoform X1 OS=Thrips palmi OX=161013 GN=LOC117654355 PE=4 SV=1
MM1 pKa = 7.46AASLNAGFNTPLGNINFGLSQSSSNANSIGLGGGGGGGVGGGFGGSTAGANAGAGGASAGGQGAGGGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGGFGGSRR129 pKa = 11.84PGGFGGPGGFGGSRR143 pKa = 11.84PGGFGGPGGFGGQGIGQSHH162 pKa = 6.05GQRR165 pKa = 11.84HH166 pKa = 4.88KK167 pKa = 10.84RR168 pKa = 11.84QFGFSGSSANAQSHH182 pKa = 5.88SGSGGFFGGGLGGLGASGANAGASSFGSGFGGLGGNLGGGLASSGANAGASSFGGGFGGLGGGLGASGANAGASSFGGGLGGLGGFGGLGGFGGFGGLGGLAGLGSGLGVSGANAGAGSFGAGGTHH308 pKa = 6.56HH309 pKa = 6.89GVGGGLSASQAASHH323 pKa = 6.32AASGAGGVGVGPTVGGGLSGSQASANAGSIGGGIGGGAGGAGGLSGSLANAQSGAGSIGGGVGLGGGVSGSQAAANAGSIGGGIGGGHH411 pKa = 6.59GFGGGLSGSQASANAGSIGGGIGGGAGGAGGLSGSLANAQSGAGSIGGGVGLGGGVTGSQAAANAGSLGGGIGGGHH487 pKa = 6.59GFGGGLSGSQASANAGSIGGGIGGGAGGAGGLSGSLANAQSGAGSIGGGVGLGGGVSGSQAAANAGSLSGGFGGGQGFGGGLSGSQASANAGSLSGGFGGGHH589 pKa = 6.39GFGGGLSGSQASANAGSLSGGFGGGHH615 pKa = 6.39GFGGGLSGSQASASAGSVAGGAGFGRR641 pKa = 4.21
MM1 pKa = 7.46AASLNAGFNTPLGNINFGLSQSSSNANSIGLGGGGGGGVGGGFGGSTAGANAGAGGASAGGQGAGGGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGFGGQGGFGGSRR129 pKa = 11.84PGGFGGPGGFGGSRR143 pKa = 11.84PGGFGGPGGFGGQGIGQSHH162 pKa = 6.05GQRR165 pKa = 11.84HH166 pKa = 4.88KK167 pKa = 10.84RR168 pKa = 11.84QFGFSGSSANAQSHH182 pKa = 5.88SGSGGFFGGGLGGLGASGANAGASSFGSGFGGLGGNLGGGLASSGANAGASSFGGGFGGLGGGLGASGANAGASSFGGGLGGLGGFGGLGGFGGFGGLGGLAGLGSGLGVSGANAGAGSFGAGGTHH308 pKa = 6.56HH309 pKa = 6.89GVGGGLSASQAASHH323 pKa = 6.32AASGAGGVGVGPTVGGGLSGSQASANAGSIGGGIGGGAGGAGGLSGSLANAQSGAGSIGGGVGLGGGVSGSQAAANAGSIGGGIGGGHH411 pKa = 6.59GFGGGLSGSQASANAGSIGGGIGGGAGGAGGLSGSLANAQSGAGSIGGGVGLGGGVTGSQAAANAGSLGGGIGGGHH487 pKa = 6.59GFGGGLSGSQASANAGSIGGGIGGGAGGAGGLSGSLANAQSGAGSIGGGVGLGGGVSGSQAAANAGSLSGGFGGGQGFGGGLSGSQASANAGSLSGGFGGGHH589 pKa = 6.39GFGGGLSGSQASANAGSLSGGFGGGHH615 pKa = 6.39GFGGGLSGSQASASAGSVAGGAGFGRR641 pKa = 4.21
Molecular weight: 53.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13368287 |
37 |
16816 |
637.5 |
70.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.87 ± 0.022 | 1.929 ± 0.018 |
5.54 ± 0.014 | 6.47 ± 0.028 |
3.297 ± 0.013 | 6.371 ± 0.021 |
2.679 ± 0.009 | 3.965 ± 0.013 |
5.49 ± 0.022 | 9.234 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.293 ± 0.01 | 3.923 ± 0.012 |
6.177 ± 0.027 | 4.619 ± 0.022 |
5.896 ± 0.019 | 8.663 ± 0.028 |
5.453 ± 0.015 | 6.592 ± 0.015 |
1.087 ± 0.005 | 2.452 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
