
Maylandia zebra (zebra mbuna)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei<
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
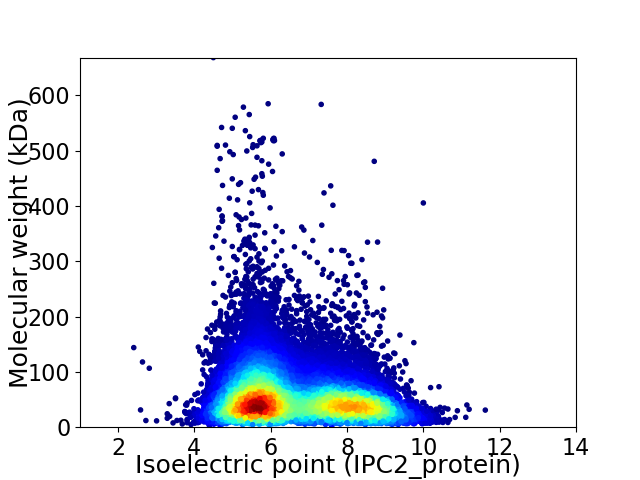
Virtual 2D-PAGE plot for 37295 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P9CSE2|A0A3P9CSE2_9CICH Nucleoporin 155 OS=Maylandia zebra OX=106582 PE=3 SV=1
MM1 pKa = 7.52LWFLGLTCVLCIGGSAVAGQRR22 pKa = 11.84DD23 pKa = 4.16GEE25 pKa = 4.35IISQIKK31 pKa = 6.94MTNLVLGEE39 pKa = 4.07IKK41 pKa = 10.42EE42 pKa = 4.1LLKK45 pKa = 10.61QQIKK49 pKa = 10.53EE50 pKa = 3.89IVFLKK55 pKa = 8.75NTVMEE60 pKa = 4.76CEE62 pKa = 4.0ACAMGGMQPQPTCVPNSCHH81 pKa = 6.45PGVSFVEE88 pKa = 4.5TPEE91 pKa = 4.08GVKK94 pKa = 10.48CGPCPHH100 pKa = 6.8GMEE103 pKa = 5.14GNGTHH108 pKa = 5.99CTDD111 pKa = 3.66VDD113 pKa = 3.7EE114 pKa = 4.91CTVKK118 pKa = 10.02PCHH121 pKa = 6.14MGVRR125 pKa = 11.84CINTSPGFRR134 pKa = 11.84CGSCPAGYY142 pKa = 8.41TGPQVQGVGLAYY154 pKa = 9.21ATANKK159 pKa = 9.17QVCRR163 pKa = 11.84DD164 pKa = 3.32INEE167 pKa = 4.25CDD169 pKa = 3.5GPNNGGCVANSVCLNTPGSFRR190 pKa = 11.84CGPCKK195 pKa = 9.95TGYY198 pKa = 8.97TGDD201 pKa = 3.59QQQGCKK207 pKa = 9.73PEE209 pKa = 4.04RR210 pKa = 11.84ACGNGQPNPCHH221 pKa = 6.71ASAEE225 pKa = 4.62CIVHH229 pKa = 6.66RR230 pKa = 11.84EE231 pKa = 4.01GTIEE235 pKa = 4.18CQCGVGWAGNGYY247 pKa = 9.89VCGPDD252 pKa = 3.17TDD254 pKa = 4.09IDD256 pKa = 3.94GFPDD260 pKa = 3.58EE261 pKa = 5.92KK262 pKa = 11.02LDD264 pKa = 5.4CPDD267 pKa = 3.83SNCNKK272 pKa = 10.03DD273 pKa = 3.32NCLTVPNSGQEE284 pKa = 4.12DD285 pKa = 4.09ADD287 pKa = 3.68SDD289 pKa = 4.74GIGDD293 pKa = 4.47ACDD296 pKa = 3.53EE297 pKa = 4.74DD298 pKa = 5.69ADD300 pKa = 4.77GDD302 pKa = 4.76GILNTQDD309 pKa = 3.01NCVLVPNVDD318 pKa = 3.12QRR320 pKa = 11.84NIDD323 pKa = 3.42EE324 pKa = 5.56DD325 pKa = 4.48DD326 pKa = 4.81FGDD329 pKa = 4.55ACDD332 pKa = 3.7NCPAVKK338 pKa = 10.8NNDD341 pKa = 3.5QKK343 pKa = 11.29DD344 pKa = 3.71TDD346 pKa = 3.32VDD348 pKa = 3.94KK349 pKa = 11.9YY350 pKa = 11.35GDD352 pKa = 3.78EE353 pKa = 4.36CDD355 pKa = 3.89EE356 pKa = 5.85DD357 pKa = 4.15IDD359 pKa = 5.21GDD361 pKa = 4.45GIPNHH366 pKa = 7.08LDD368 pKa = 2.97NCKK371 pKa = 9.84RR372 pKa = 11.84VPNADD377 pKa = 3.2QKK379 pKa = 11.71DD380 pKa = 3.63RR381 pKa = 11.84DD382 pKa = 3.66GDD384 pKa = 4.12KK385 pKa = 11.46VGDD388 pKa = 4.05ACDD391 pKa = 3.46SCPYY395 pKa = 10.53DD396 pKa = 3.99PNPDD400 pKa = 3.95QMDD403 pKa = 4.12ADD405 pKa = 4.07NDD407 pKa = 4.82LIGDD411 pKa = 3.97PCDD414 pKa = 3.62TNKK417 pKa = 10.93DD418 pKa = 3.49SDD420 pKa = 4.16GDD422 pKa = 3.81GHH424 pKa = 6.91QDD426 pKa = 4.2SRR428 pKa = 11.84DD429 pKa = 3.47NCPAVINSSQLDD441 pKa = 3.5TDD443 pKa = 3.78KK444 pKa = 11.37DD445 pKa = 4.07GKK447 pKa = 11.12GDD449 pKa = 3.71EE450 pKa = 5.3CDD452 pKa = 5.61DD453 pKa = 5.63DD454 pKa = 6.35DD455 pKa = 7.03DD456 pKa = 6.2NDD458 pKa = 5.44GIPDD462 pKa = 4.89LLPPGPDD469 pKa = 2.97NCRR472 pKa = 11.84LIPNPLQEE480 pKa = 5.54DD481 pKa = 3.65SDD483 pKa = 4.45GDD485 pKa = 4.06GVGNVCEE492 pKa = 4.73KK493 pKa = 11.11DD494 pKa = 3.31FDD496 pKa = 4.13NDD498 pKa = 4.31TIIDD502 pKa = 4.92PIDD505 pKa = 3.6VCPEE509 pKa = 3.74NAEE512 pKa = 4.04VTLTDD517 pKa = 3.59FRR519 pKa = 11.84EE520 pKa = 4.27YY521 pKa = 9.18QTVVLDD527 pKa = 4.15PEE529 pKa = 5.32GDD531 pKa = 3.65AQIDD535 pKa = 4.13PNWVVLNQGRR545 pKa = 11.84EE546 pKa = 3.9IVQTMNSDD554 pKa = 3.19PGLAVGYY561 pKa = 7.12TAFSGVDD568 pKa = 3.72FEE570 pKa = 5.24GTFHH574 pKa = 6.81VNTVTDD580 pKa = 3.57DD581 pKa = 4.08DD582 pKa = 4.34YY583 pKa = 12.0AGFIFAYY590 pKa = 9.51QDD592 pKa = 3.06SSSFYY597 pKa = 9.91VVMWKK602 pKa = 9.52QVEE605 pKa = 4.18QIYY608 pKa = 8.33WQANPFRR615 pKa = 11.84AVAEE619 pKa = 4.08PGIQLKK625 pKa = 10.06AVKK628 pKa = 10.35SNTGPGEE635 pKa = 4.06NLRR638 pKa = 11.84NALWHH643 pKa = 6.46TGDD646 pKa = 3.25TTDD649 pKa = 4.87QVKK652 pKa = 10.44LLWKK656 pKa = 10.05DD657 pKa = 3.18ARR659 pKa = 11.84NVGWKK664 pKa = 10.41DD665 pKa = 3.0KK666 pKa = 10.83TSYY669 pKa = 10.52RR670 pKa = 11.84WFLQHH675 pKa = 6.87RR676 pKa = 11.84PADD679 pKa = 3.73GYY681 pKa = 10.45IRR683 pKa = 11.84VRR685 pKa = 11.84FYY687 pKa = 11.29EE688 pKa = 4.41GTQMVADD695 pKa = 3.86TGVIIDD701 pKa = 3.35ATMRR705 pKa = 11.84GGRR708 pKa = 11.84LGVFCFSQEE717 pKa = 3.95NIIWANLRR725 pKa = 11.84YY726 pKa = 9.71RR727 pKa = 11.84CNDD730 pKa = 3.48TLPEE734 pKa = 4.48DD735 pKa = 3.93FDD737 pKa = 4.54AYY739 pKa = 10.45RR740 pKa = 11.84AQQVQLVAA748 pKa = 4.4
MM1 pKa = 7.52LWFLGLTCVLCIGGSAVAGQRR22 pKa = 11.84DD23 pKa = 4.16GEE25 pKa = 4.35IISQIKK31 pKa = 6.94MTNLVLGEE39 pKa = 4.07IKK41 pKa = 10.42EE42 pKa = 4.1LLKK45 pKa = 10.61QQIKK49 pKa = 10.53EE50 pKa = 3.89IVFLKK55 pKa = 8.75NTVMEE60 pKa = 4.76CEE62 pKa = 4.0ACAMGGMQPQPTCVPNSCHH81 pKa = 6.45PGVSFVEE88 pKa = 4.5TPEE91 pKa = 4.08GVKK94 pKa = 10.48CGPCPHH100 pKa = 6.8GMEE103 pKa = 5.14GNGTHH108 pKa = 5.99CTDD111 pKa = 3.66VDD113 pKa = 3.7EE114 pKa = 4.91CTVKK118 pKa = 10.02PCHH121 pKa = 6.14MGVRR125 pKa = 11.84CINTSPGFRR134 pKa = 11.84CGSCPAGYY142 pKa = 8.41TGPQVQGVGLAYY154 pKa = 9.21ATANKK159 pKa = 9.17QVCRR163 pKa = 11.84DD164 pKa = 3.32INEE167 pKa = 4.25CDD169 pKa = 3.5GPNNGGCVANSVCLNTPGSFRR190 pKa = 11.84CGPCKK195 pKa = 9.95TGYY198 pKa = 8.97TGDD201 pKa = 3.59QQQGCKK207 pKa = 9.73PEE209 pKa = 4.04RR210 pKa = 11.84ACGNGQPNPCHH221 pKa = 6.71ASAEE225 pKa = 4.62CIVHH229 pKa = 6.66RR230 pKa = 11.84EE231 pKa = 4.01GTIEE235 pKa = 4.18CQCGVGWAGNGYY247 pKa = 9.89VCGPDD252 pKa = 3.17TDD254 pKa = 4.09IDD256 pKa = 3.94GFPDD260 pKa = 3.58EE261 pKa = 5.92KK262 pKa = 11.02LDD264 pKa = 5.4CPDD267 pKa = 3.83SNCNKK272 pKa = 10.03DD273 pKa = 3.32NCLTVPNSGQEE284 pKa = 4.12DD285 pKa = 4.09ADD287 pKa = 3.68SDD289 pKa = 4.74GIGDD293 pKa = 4.47ACDD296 pKa = 3.53EE297 pKa = 4.74DD298 pKa = 5.69ADD300 pKa = 4.77GDD302 pKa = 4.76GILNTQDD309 pKa = 3.01NCVLVPNVDD318 pKa = 3.12QRR320 pKa = 11.84NIDD323 pKa = 3.42EE324 pKa = 5.56DD325 pKa = 4.48DD326 pKa = 4.81FGDD329 pKa = 4.55ACDD332 pKa = 3.7NCPAVKK338 pKa = 10.8NNDD341 pKa = 3.5QKK343 pKa = 11.29DD344 pKa = 3.71TDD346 pKa = 3.32VDD348 pKa = 3.94KK349 pKa = 11.9YY350 pKa = 11.35GDD352 pKa = 3.78EE353 pKa = 4.36CDD355 pKa = 3.89EE356 pKa = 5.85DD357 pKa = 4.15IDD359 pKa = 5.21GDD361 pKa = 4.45GIPNHH366 pKa = 7.08LDD368 pKa = 2.97NCKK371 pKa = 9.84RR372 pKa = 11.84VPNADD377 pKa = 3.2QKK379 pKa = 11.71DD380 pKa = 3.63RR381 pKa = 11.84DD382 pKa = 3.66GDD384 pKa = 4.12KK385 pKa = 11.46VGDD388 pKa = 4.05ACDD391 pKa = 3.46SCPYY395 pKa = 10.53DD396 pKa = 3.99PNPDD400 pKa = 3.95QMDD403 pKa = 4.12ADD405 pKa = 4.07NDD407 pKa = 4.82LIGDD411 pKa = 3.97PCDD414 pKa = 3.62TNKK417 pKa = 10.93DD418 pKa = 3.49SDD420 pKa = 4.16GDD422 pKa = 3.81GHH424 pKa = 6.91QDD426 pKa = 4.2SRR428 pKa = 11.84DD429 pKa = 3.47NCPAVINSSQLDD441 pKa = 3.5TDD443 pKa = 3.78KK444 pKa = 11.37DD445 pKa = 4.07GKK447 pKa = 11.12GDD449 pKa = 3.71EE450 pKa = 5.3CDD452 pKa = 5.61DD453 pKa = 5.63DD454 pKa = 6.35DD455 pKa = 7.03DD456 pKa = 6.2NDD458 pKa = 5.44GIPDD462 pKa = 4.89LLPPGPDD469 pKa = 2.97NCRR472 pKa = 11.84LIPNPLQEE480 pKa = 5.54DD481 pKa = 3.65SDD483 pKa = 4.45GDD485 pKa = 4.06GVGNVCEE492 pKa = 4.73KK493 pKa = 11.11DD494 pKa = 3.31FDD496 pKa = 4.13NDD498 pKa = 4.31TIIDD502 pKa = 4.92PIDD505 pKa = 3.6VCPEE509 pKa = 3.74NAEE512 pKa = 4.04VTLTDD517 pKa = 3.59FRR519 pKa = 11.84EE520 pKa = 4.27YY521 pKa = 9.18QTVVLDD527 pKa = 4.15PEE529 pKa = 5.32GDD531 pKa = 3.65AQIDD535 pKa = 4.13PNWVVLNQGRR545 pKa = 11.84EE546 pKa = 3.9IVQTMNSDD554 pKa = 3.19PGLAVGYY561 pKa = 7.12TAFSGVDD568 pKa = 3.72FEE570 pKa = 5.24GTFHH574 pKa = 6.81VNTVTDD580 pKa = 3.57DD581 pKa = 4.08DD582 pKa = 4.34YY583 pKa = 12.0AGFIFAYY590 pKa = 9.51QDD592 pKa = 3.06SSSFYY597 pKa = 9.91VVMWKK602 pKa = 9.52QVEE605 pKa = 4.18QIYY608 pKa = 8.33WQANPFRR615 pKa = 11.84AVAEE619 pKa = 4.08PGIQLKK625 pKa = 10.06AVKK628 pKa = 10.35SNTGPGEE635 pKa = 4.06NLRR638 pKa = 11.84NALWHH643 pKa = 6.46TGDD646 pKa = 3.25TTDD649 pKa = 4.87QVKK652 pKa = 10.44LLWKK656 pKa = 10.05DD657 pKa = 3.18ARR659 pKa = 11.84NVGWKK664 pKa = 10.41DD665 pKa = 3.0KK666 pKa = 10.83TSYY669 pKa = 10.52RR670 pKa = 11.84WFLQHH675 pKa = 6.87RR676 pKa = 11.84PADD679 pKa = 3.73GYY681 pKa = 10.45IRR683 pKa = 11.84VRR685 pKa = 11.84FYY687 pKa = 11.29EE688 pKa = 4.41GTQMVADD695 pKa = 3.86TGVIIDD701 pKa = 3.35ATMRR705 pKa = 11.84GGRR708 pKa = 11.84LGVFCFSQEE717 pKa = 3.95NIIWANLRR725 pKa = 11.84YY726 pKa = 9.71RR727 pKa = 11.84CNDD730 pKa = 3.48TLPEE734 pKa = 4.48DD735 pKa = 3.93FDD737 pKa = 4.54AYY739 pKa = 10.45RR740 pKa = 11.84AQQVQLVAA748 pKa = 4.4
Molecular weight: 81.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P9CG60|A0A3P9CG60_9CICH Ankyrin repeat and kinase domain containing 1 OS=Maylandia zebra OX=106582 PE=4 SV=1
MM1 pKa = 7.55CIVSGVLAIRR11 pKa = 11.84SLSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPPSVQSPVSPLAQPLTPPQAPFSPLPSLQSSVRR191 pKa = 11.84SATQSPLAPSSTLPPVSLPPQPSLHH216 pKa = 7.0PATQAATQSVIQPILRR232 pKa = 11.84LARR235 pKa = 11.84PQPLRR240 pKa = 11.84PRR242 pKa = 11.84LARR245 pKa = 11.84PPQPLRR251 pKa = 11.84PRR253 pKa = 11.84LARR256 pKa = 11.84PPQPLRR262 pKa = 11.84PRR264 pKa = 11.84LARR267 pKa = 11.84PPQSLRR273 pKa = 11.84PRR275 pKa = 11.84LARR278 pKa = 11.84PPQSLRR284 pKa = 11.84PRR286 pKa = 11.84LARR289 pKa = 11.84PPQSLRR295 pKa = 11.84LLQPQPVLRR304 pKa = 11.84LVPPP308 pKa = 4.66
MM1 pKa = 7.55CIVSGVLAIRR11 pKa = 11.84SLSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAQSAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPFTAPLAPSQPPSVQSPVSPLAQPLTPPQAPFSPLPSLQSSVRR191 pKa = 11.84SATQSPLAPSSTLPPVSLPPQPSLHH216 pKa = 7.0PATQAATQSVIQPILRR232 pKa = 11.84LARR235 pKa = 11.84PQPLRR240 pKa = 11.84PRR242 pKa = 11.84LARR245 pKa = 11.84PPQPLRR251 pKa = 11.84PRR253 pKa = 11.84LARR256 pKa = 11.84PPQPLRR262 pKa = 11.84PRR264 pKa = 11.84LARR267 pKa = 11.84PPQSLRR273 pKa = 11.84PRR275 pKa = 11.84LARR278 pKa = 11.84PPQSLRR284 pKa = 11.84PRR286 pKa = 11.84LARR289 pKa = 11.84PPQSLRR295 pKa = 11.84LLQPQPVLRR304 pKa = 11.84LVPPP308 pKa = 4.66
Molecular weight: 32.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
20228919 |
17 |
6169 |
542.4 |
60.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.417 ± 0.01 | 2.329 ± 0.01 |
5.135 ± 0.009 | 6.74 ± 0.016 |
3.75 ± 0.009 | 6.202 ± 0.016 |
2.658 ± 0.006 | 4.574 ± 0.01 |
5.766 ± 0.015 | 9.552 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.342 ± 0.005 | 3.951 ± 0.008 |
5.55 ± 0.017 | 4.667 ± 0.012 |
5.494 ± 0.012 | 8.739 ± 0.016 |
5.751 ± 0.01 | 6.38 ± 0.01 |
1.174 ± 0.005 | 2.826 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
