
Xylaria flabelliformis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Xylariaceae; Xylaria
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
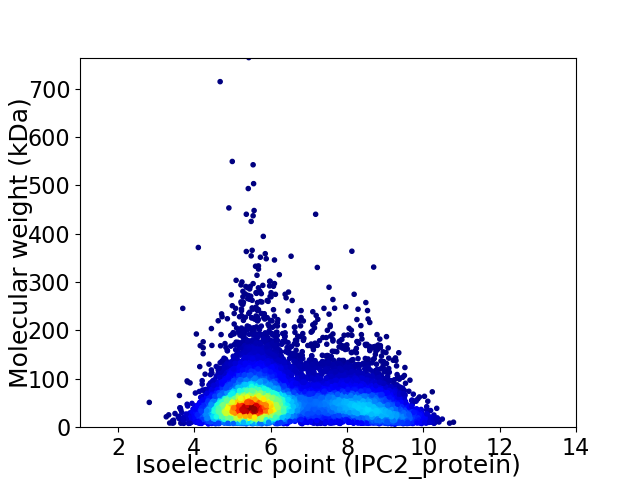
Virtual 2D-PAGE plot for 11403 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
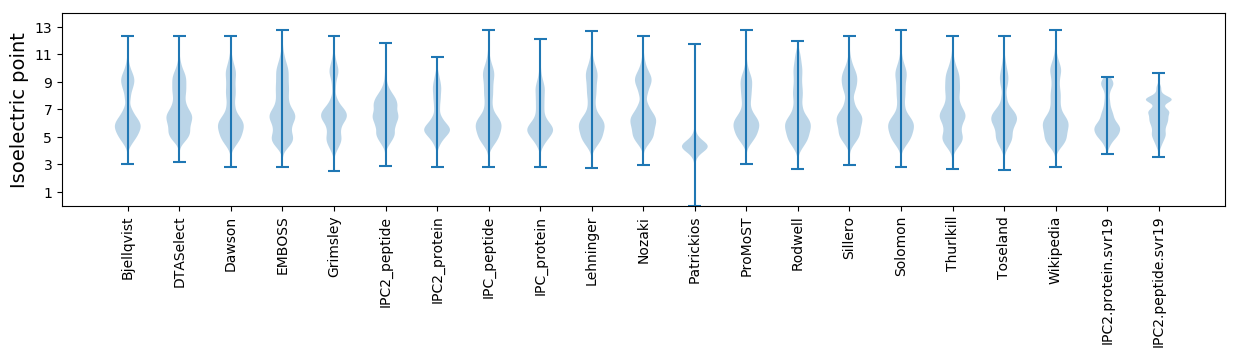
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553HZZ0|A0A553HZZ0_9PEZI M20_dimer domain-containing protein OS=Xylaria flabelliformis OX=2512241 GN=FHL15_005492 PE=3 SV=1
MM1 pKa = 7.24PLKK4 pKa = 10.81YY5 pKa = 11.15NMMDD9 pKa = 3.82LCRR12 pKa = 11.84SDD14 pKa = 3.28NLRR17 pKa = 11.84YY18 pKa = 9.87VYY20 pKa = 10.31YY21 pKa = 10.72GQIWWDD27 pKa = 3.51NDD29 pKa = 3.23SDD31 pKa = 4.52PDD33 pKa = 4.06NLYY36 pKa = 11.11DD37 pKa = 6.55LDD39 pKa = 5.77DD40 pKa = 6.03LDD42 pKa = 6.35DD43 pKa = 5.74PDD45 pKa = 6.08DD46 pKa = 4.73DD47 pKa = 5.89GYY49 pKa = 11.04INSSDD54 pKa = 4.42KK55 pKa = 11.2EE56 pKa = 4.22DD57 pKa = 3.68ANKK60 pKa = 10.54DD61 pKa = 3.34AEE63 pKa = 4.48KK64 pKa = 10.96DD65 pKa = 3.5NKK67 pKa = 10.3FYY69 pKa = 11.44
MM1 pKa = 7.24PLKK4 pKa = 10.81YY5 pKa = 11.15NMMDD9 pKa = 3.82LCRR12 pKa = 11.84SDD14 pKa = 3.28NLRR17 pKa = 11.84YY18 pKa = 9.87VYY20 pKa = 10.31YY21 pKa = 10.72GQIWWDD27 pKa = 3.51NDD29 pKa = 3.23SDD31 pKa = 4.52PDD33 pKa = 4.06NLYY36 pKa = 11.11DD37 pKa = 6.55LDD39 pKa = 5.77DD40 pKa = 6.03LDD42 pKa = 6.35DD43 pKa = 5.74PDD45 pKa = 6.08DD46 pKa = 4.73DD47 pKa = 5.89GYY49 pKa = 11.04INSSDD54 pKa = 4.42KK55 pKa = 11.2EE56 pKa = 4.22DD57 pKa = 3.68ANKK60 pKa = 10.54DD61 pKa = 3.34AEE63 pKa = 4.48KK64 pKa = 10.96DD65 pKa = 3.5NKK67 pKa = 10.3FYY69 pKa = 11.44
Molecular weight: 8.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553HYA3|A0A553HYA3_9PEZI Uncharacterized protein OS=Xylaria flabelliformis OX=2512241 GN=FHL15_006067 PE=4 SV=1
MM1 pKa = 7.31TSSTPPLLQRR11 pKa = 11.84PRR13 pKa = 11.84AHH15 pKa = 7.29HH16 pKa = 6.56ARR18 pKa = 11.84LDD20 pKa = 3.7KK21 pKa = 10.61TIPPALRR28 pKa = 11.84PVIRR32 pKa = 11.84AYY34 pKa = 10.9VLGYY38 pKa = 10.6ASAVAPRR45 pKa = 11.84VLTLVLQYY53 pKa = 10.12GARR56 pKa = 11.84WRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84KK61 pKa = 10.02SRR63 pKa = 11.84GPVIVNGTQPQDD75 pKa = 3.26SFVASLRR82 pKa = 11.84RR83 pKa = 11.84ILSGGLDD90 pKa = 3.52PQRR93 pKa = 11.84FPTFCATLIGGTTLLDD109 pKa = 3.44VLVRR113 pKa = 11.84RR114 pKa = 11.84LMKK117 pKa = 10.09RR118 pKa = 11.84WKK120 pKa = 8.62PQLSGIVRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84LSIWISSFIAGWLSLRR146 pKa = 11.84LLQSKK151 pKa = 8.0KK152 pKa = 8.58TNAFAEE158 pKa = 4.78TKK160 pKa = 10.33AVNDD164 pKa = 4.51DD165 pKa = 3.39KK166 pKa = 11.61QKK168 pKa = 8.63TIRR171 pKa = 11.84YY172 pKa = 8.75AGRR175 pKa = 11.84TLDD178 pKa = 3.65LTLFAATRR186 pKa = 11.84AVDD189 pKa = 3.8VVVGEE194 pKa = 4.52LWSQRR199 pKa = 11.84RR200 pKa = 11.84QRR202 pKa = 11.84RR203 pKa = 11.84VAADD207 pKa = 2.97QWSWFDD213 pKa = 3.65SFISKK218 pKa = 9.03MADD221 pKa = 3.01PSIFATSCALIMWAWFYY238 pKa = 10.07TPCALPKK245 pKa = 10.33SYY247 pKa = 11.31NKK249 pKa = 9.45WISSAAAVDD258 pKa = 3.61SRR260 pKa = 11.84LIEE263 pKa = 4.0ALRR266 pKa = 11.84RR267 pKa = 11.84CHH269 pKa = 7.1KK270 pKa = 10.93GEE272 pKa = 3.76LRR274 pKa = 11.84YY275 pKa = 10.49GEE277 pKa = 4.22EE278 pKa = 3.97TGQAPLLQSMCSDD291 pKa = 3.73YY292 pKa = 11.04KK293 pKa = 10.81WPLEE297 pKa = 4.12WGDD300 pKa = 3.63PATTVPFPCEE310 pKa = 3.72IVHH313 pKa = 6.09MGCGPSCEE321 pKa = 3.93YY322 pKa = 10.24HH323 pKa = 5.61ALYY326 pKa = 10.66RR327 pKa = 11.84FFRR330 pKa = 11.84SFKK333 pKa = 10.01WSMATYY339 pKa = 10.7LPLNLLARR347 pKa = 11.84KK348 pKa = 9.47KK349 pKa = 10.36DD350 pKa = 3.47LQGFRR355 pKa = 11.84KK356 pKa = 9.98ALTSAARR363 pKa = 11.84SSSFLATFITLFYY376 pKa = 10.98YY377 pKa = 8.72GVCLARR383 pKa = 11.84TRR385 pKa = 11.84LGPRR389 pKa = 11.84LLGKK393 pKa = 8.49HH394 pKa = 4.43TASRR398 pKa = 11.84QKK400 pKa = 10.09IDD402 pKa = 3.35AGVCVGCGCFMCGWSIFFEE421 pKa = 4.25AASRR425 pKa = 11.84RR426 pKa = 11.84KK427 pKa = 10.55DD428 pKa = 3.1MALFVAPRR436 pKa = 11.84ALATLLPRR444 pKa = 11.84RR445 pKa = 11.84YY446 pKa = 9.14PLNKK450 pKa = 8.59QWRR453 pKa = 11.84EE454 pKa = 3.67TFVFAFSTAIVFTCVRR470 pKa = 11.84EE471 pKa = 3.69NRR473 pKa = 11.84ARR475 pKa = 11.84VRR477 pKa = 11.84GVLGNILGSVLEE489 pKa = 4.41TT490 pKa = 3.81
MM1 pKa = 7.31TSSTPPLLQRR11 pKa = 11.84PRR13 pKa = 11.84AHH15 pKa = 7.29HH16 pKa = 6.56ARR18 pKa = 11.84LDD20 pKa = 3.7KK21 pKa = 10.61TIPPALRR28 pKa = 11.84PVIRR32 pKa = 11.84AYY34 pKa = 10.9VLGYY38 pKa = 10.6ASAVAPRR45 pKa = 11.84VLTLVLQYY53 pKa = 10.12GARR56 pKa = 11.84WRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84KK61 pKa = 10.02SRR63 pKa = 11.84GPVIVNGTQPQDD75 pKa = 3.26SFVASLRR82 pKa = 11.84RR83 pKa = 11.84ILSGGLDD90 pKa = 3.52PQRR93 pKa = 11.84FPTFCATLIGGTTLLDD109 pKa = 3.44VLVRR113 pKa = 11.84RR114 pKa = 11.84LMKK117 pKa = 10.09RR118 pKa = 11.84WKK120 pKa = 8.62PQLSGIVRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84LSIWISSFIAGWLSLRR146 pKa = 11.84LLQSKK151 pKa = 8.0KK152 pKa = 8.58TNAFAEE158 pKa = 4.78TKK160 pKa = 10.33AVNDD164 pKa = 4.51DD165 pKa = 3.39KK166 pKa = 11.61QKK168 pKa = 8.63TIRR171 pKa = 11.84YY172 pKa = 8.75AGRR175 pKa = 11.84TLDD178 pKa = 3.65LTLFAATRR186 pKa = 11.84AVDD189 pKa = 3.8VVVGEE194 pKa = 4.52LWSQRR199 pKa = 11.84RR200 pKa = 11.84QRR202 pKa = 11.84RR203 pKa = 11.84VAADD207 pKa = 2.97QWSWFDD213 pKa = 3.65SFISKK218 pKa = 9.03MADD221 pKa = 3.01PSIFATSCALIMWAWFYY238 pKa = 10.07TPCALPKK245 pKa = 10.33SYY247 pKa = 11.31NKK249 pKa = 9.45WISSAAAVDD258 pKa = 3.61SRR260 pKa = 11.84LIEE263 pKa = 4.0ALRR266 pKa = 11.84RR267 pKa = 11.84CHH269 pKa = 7.1KK270 pKa = 10.93GEE272 pKa = 3.76LRR274 pKa = 11.84YY275 pKa = 10.49GEE277 pKa = 4.22EE278 pKa = 3.97TGQAPLLQSMCSDD291 pKa = 3.73YY292 pKa = 11.04KK293 pKa = 10.81WPLEE297 pKa = 4.12WGDD300 pKa = 3.63PATTVPFPCEE310 pKa = 3.72IVHH313 pKa = 6.09MGCGPSCEE321 pKa = 3.93YY322 pKa = 10.24HH323 pKa = 5.61ALYY326 pKa = 10.66RR327 pKa = 11.84FFRR330 pKa = 11.84SFKK333 pKa = 10.01WSMATYY339 pKa = 10.7LPLNLLARR347 pKa = 11.84KK348 pKa = 9.47KK349 pKa = 10.36DD350 pKa = 3.47LQGFRR355 pKa = 11.84KK356 pKa = 9.98ALTSAARR363 pKa = 11.84SSSFLATFITLFYY376 pKa = 10.98YY377 pKa = 8.72GVCLARR383 pKa = 11.84TRR385 pKa = 11.84LGPRR389 pKa = 11.84LLGKK393 pKa = 8.49HH394 pKa = 4.43TASRR398 pKa = 11.84QKK400 pKa = 10.09IDD402 pKa = 3.35AGVCVGCGCFMCGWSIFFEE421 pKa = 4.25AASRR425 pKa = 11.84RR426 pKa = 11.84KK427 pKa = 10.55DD428 pKa = 3.1MALFVAPRR436 pKa = 11.84ALATLLPRR444 pKa = 11.84RR445 pKa = 11.84YY446 pKa = 9.14PLNKK450 pKa = 8.59QWRR453 pKa = 11.84EE454 pKa = 3.67TFVFAFSTAIVFTCVRR470 pKa = 11.84EE471 pKa = 3.69NRR473 pKa = 11.84ARR475 pKa = 11.84VRR477 pKa = 11.84GVLGNILGSVLEE489 pKa = 4.41TT490 pKa = 3.81
Molecular weight: 55.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5968531 |
57 |
6921 |
523.4 |
57.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.491 ± 0.02 | 1.207 ± 0.007 |
5.83 ± 0.015 | 6.072 ± 0.02 |
3.668 ± 0.013 | 6.744 ± 0.02 |
2.416 ± 0.009 | 5.097 ± 0.015 |
4.774 ± 0.015 | 8.893 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.069 ± 0.007 | 3.848 ± 0.012 |
5.981 ± 0.023 | 3.971 ± 0.014 |
6.146 ± 0.02 | 8.292 ± 0.025 |
6.137 ± 0.017 | 6.069 ± 0.016 |
1.472 ± 0.008 | 2.821 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
