
Flavobacterium sp. FPG59
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
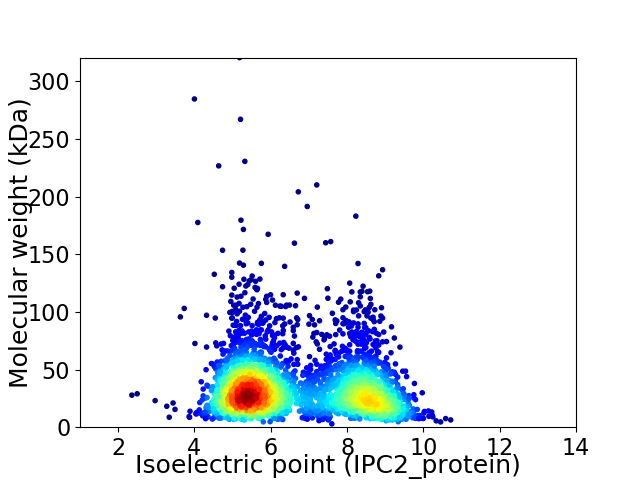
Virtual 2D-PAGE plot for 3173 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A243SC85|A0A243SC85_9FLAO Uncharacterized protein (Fragment) OS=Flavobacterium sp. FPG59 OX=1929267 GN=FPG59_15490 PE=4 SV=1
MM1 pKa = 7.6NIQLPSFIFKK11 pKa = 10.39VPRR14 pKa = 11.84FLFLFWSLYY23 pKa = 7.75TVNSSAQVTANTNPSNAQINLALQGPGIVITGGTLVGAQTDD64 pKa = 4.35GAILRR69 pKa = 11.84SNQVATFTNGITGAGLGFPSGAYY92 pKa = 9.93FSTGNTTLEE101 pKa = 3.94LTNRR105 pKa = 11.84NTVIQQSAFPAAGATLSDD123 pKa = 4.44ADD125 pKa = 5.11LSTIDD130 pKa = 3.46PTATRR135 pKa = 11.84DD136 pKa = 3.32LVAYY140 pKa = 7.36TFTITLGPTITGLRR154 pKa = 11.84IGYY157 pKa = 9.14HH158 pKa = 6.12FGSEE162 pKa = 4.36EE163 pKa = 3.97YY164 pKa = 9.22PDD166 pKa = 3.74YY167 pKa = 11.35VGTVFDD173 pKa = 4.44DD174 pKa = 3.69AFGFFISGPGITGTRR189 pKa = 11.84NLATTPAGSPTSINKK204 pKa = 9.54INGGFPGSVAGATPVAAYY222 pKa = 10.2DD223 pKa = 3.87GTQSALYY230 pKa = 9.43INNGHH235 pKa = 5.43TTTVSGGAYY244 pKa = 9.25VANTDD249 pKa = 3.83PQPGPFPIFTEE260 pKa = 4.57FNGVTRR266 pKa = 11.84FITRR270 pKa = 11.84TVNNLTPGGTYY281 pKa = 7.71TFKK284 pKa = 11.21VVIADD289 pKa = 3.75ASDD292 pKa = 4.05DD293 pKa = 3.89FLDD296 pKa = 4.28SGVFLNLIEE305 pKa = 4.66GLTNANLQISKK316 pKa = 8.65TVNNPTPIVGSNVTFTLTASNLGSGAATGVNVTDD350 pKa = 4.61ILPAGYY356 pKa = 7.92TFDD359 pKa = 3.55SALPSVGTYY368 pKa = 11.24DD369 pKa = 3.25NTTGLWSIGDD379 pKa = 3.85LANGGNATLSIVARR393 pKa = 11.84VNASASYY400 pKa = 10.92INTATIAGNEE410 pKa = 4.01TDD412 pKa = 4.48PVLGNNTSTASIAIQPDD429 pKa = 3.32SDD431 pKa = 3.79NDD433 pKa = 4.02GVGDD437 pKa = 4.28FTDD440 pKa = 5.44LDD442 pKa = 4.45DD443 pKa = 6.1DD444 pKa = 4.25NDD446 pKa = 4.7GIRR449 pKa = 11.84DD450 pKa = 3.51IVEE453 pKa = 4.24DD454 pKa = 4.27TNCASGPFVVEE465 pKa = 4.29NILNEE470 pKa = 4.06VFEE473 pKa = 4.57TGTVGFPVPKK483 pKa = 9.83GDD485 pKa = 3.56LVGVPGIVGLLEE497 pKa = 4.87AYY499 pKa = 10.18NSNPGNSSTTNVGEE513 pKa = 4.09YY514 pKa = 10.4SSVVGFDD521 pKa = 3.42GNLTKK526 pKa = 10.7VINANGGGIVDD537 pKa = 4.01QLQMSSFVVKK547 pKa = 10.75KK548 pKa = 10.78NIDD551 pKa = 3.64LLAGNIITVSADD563 pKa = 3.22FSLDD567 pKa = 3.19MAAGGPSNEE576 pKa = 3.92YY577 pKa = 10.78GIALGAGGQDD587 pKa = 3.95PIWRR591 pKa = 11.84DD592 pKa = 3.74DD593 pKa = 3.66VPGVPDD599 pKa = 3.85GVFLYY604 pKa = 10.46GHH606 pKa = 6.49GVSLIRR612 pKa = 11.84EE613 pKa = 4.39PNSSGPTFTAPSRR626 pKa = 11.84VTGWFRR632 pKa = 11.84QKK634 pKa = 9.32STYY637 pKa = 10.23YY638 pKa = 10.05VANNGSGVLHH648 pKa = 7.0LYY650 pKa = 10.68ANNEE654 pKa = 4.09GYY656 pKa = 10.4QYY658 pKa = 11.13GPTGIPTSAPIIANGINFGPASNYY682 pKa = 8.76PWLNNASLSVSVDD695 pKa = 3.22QYY697 pKa = 11.44VDD699 pKa = 3.57NIVINVNHH707 pKa = 7.26CDD709 pKa = 3.62FDD711 pKa = 5.97GDD713 pKa = 4.17GVQNSFDD720 pKa = 4.54LDD722 pKa = 3.86SDD724 pKa = 3.87NDD726 pKa = 3.96GCSDD730 pKa = 3.79SNEE733 pKa = 4.29YY734 pKa = 10.33YY735 pKa = 11.34NNATSAASGQQFGQTGGAVAPVNANGTVNLPAATYY770 pKa = 8.39TGSYY774 pKa = 11.06ANATSVGTSSSIATQPVNRR793 pKa = 11.84IINAGTNTTFSVAAAGGSGVTQYY816 pKa = 9.94QWQVDD821 pKa = 4.01SGSGFTDD828 pKa = 3.47ISNGAVYY835 pKa = 11.1NNATTNTLTLTSVPATFNGYY855 pKa = 7.57VYY857 pKa = 10.43RR858 pKa = 11.84VIIRR862 pKa = 11.84EE863 pKa = 4.31SNFVCSSVVSAEE875 pKa = 3.81RR876 pKa = 11.84TLTVFPAPVVTIADD890 pKa = 3.52ASITEE895 pKa = 4.23GGNLSFPVTLSNPSATDD912 pKa = 3.01ITVTLGFTNVTTANGDD928 pKa = 3.71YY929 pKa = 7.76TTVPVTVTFLAGATTATATVPTTADD954 pKa = 3.55TIDD957 pKa = 3.89EE958 pKa = 4.37LDD960 pKa = 3.4EE961 pKa = 4.48TFTVAITSTTGTVGSTTDD979 pKa = 3.24TATGTINDD987 pKa = 4.83DD988 pKa = 3.86DD989 pKa = 4.29NAPTVASISPSNAA1002 pKa = 2.79
MM1 pKa = 7.6NIQLPSFIFKK11 pKa = 10.39VPRR14 pKa = 11.84FLFLFWSLYY23 pKa = 7.75TVNSSAQVTANTNPSNAQINLALQGPGIVITGGTLVGAQTDD64 pKa = 4.35GAILRR69 pKa = 11.84SNQVATFTNGITGAGLGFPSGAYY92 pKa = 9.93FSTGNTTLEE101 pKa = 3.94LTNRR105 pKa = 11.84NTVIQQSAFPAAGATLSDD123 pKa = 4.44ADD125 pKa = 5.11LSTIDD130 pKa = 3.46PTATRR135 pKa = 11.84DD136 pKa = 3.32LVAYY140 pKa = 7.36TFTITLGPTITGLRR154 pKa = 11.84IGYY157 pKa = 9.14HH158 pKa = 6.12FGSEE162 pKa = 4.36EE163 pKa = 3.97YY164 pKa = 9.22PDD166 pKa = 3.74YY167 pKa = 11.35VGTVFDD173 pKa = 4.44DD174 pKa = 3.69AFGFFISGPGITGTRR189 pKa = 11.84NLATTPAGSPTSINKK204 pKa = 9.54INGGFPGSVAGATPVAAYY222 pKa = 10.2DD223 pKa = 3.87GTQSALYY230 pKa = 9.43INNGHH235 pKa = 5.43TTTVSGGAYY244 pKa = 9.25VANTDD249 pKa = 3.83PQPGPFPIFTEE260 pKa = 4.57FNGVTRR266 pKa = 11.84FITRR270 pKa = 11.84TVNNLTPGGTYY281 pKa = 7.71TFKK284 pKa = 11.21VVIADD289 pKa = 3.75ASDD292 pKa = 4.05DD293 pKa = 3.89FLDD296 pKa = 4.28SGVFLNLIEE305 pKa = 4.66GLTNANLQISKK316 pKa = 8.65TVNNPTPIVGSNVTFTLTASNLGSGAATGVNVTDD350 pKa = 4.61ILPAGYY356 pKa = 7.92TFDD359 pKa = 3.55SALPSVGTYY368 pKa = 11.24DD369 pKa = 3.25NTTGLWSIGDD379 pKa = 3.85LANGGNATLSIVARR393 pKa = 11.84VNASASYY400 pKa = 10.92INTATIAGNEE410 pKa = 4.01TDD412 pKa = 4.48PVLGNNTSTASIAIQPDD429 pKa = 3.32SDD431 pKa = 3.79NDD433 pKa = 4.02GVGDD437 pKa = 4.28FTDD440 pKa = 5.44LDD442 pKa = 4.45DD443 pKa = 6.1DD444 pKa = 4.25NDD446 pKa = 4.7GIRR449 pKa = 11.84DD450 pKa = 3.51IVEE453 pKa = 4.24DD454 pKa = 4.27TNCASGPFVVEE465 pKa = 4.29NILNEE470 pKa = 4.06VFEE473 pKa = 4.57TGTVGFPVPKK483 pKa = 9.83GDD485 pKa = 3.56LVGVPGIVGLLEE497 pKa = 4.87AYY499 pKa = 10.18NSNPGNSSTTNVGEE513 pKa = 4.09YY514 pKa = 10.4SSVVGFDD521 pKa = 3.42GNLTKK526 pKa = 10.7VINANGGGIVDD537 pKa = 4.01QLQMSSFVVKK547 pKa = 10.75KK548 pKa = 10.78NIDD551 pKa = 3.64LLAGNIITVSADD563 pKa = 3.22FSLDD567 pKa = 3.19MAAGGPSNEE576 pKa = 3.92YY577 pKa = 10.78GIALGAGGQDD587 pKa = 3.95PIWRR591 pKa = 11.84DD592 pKa = 3.74DD593 pKa = 3.66VPGVPDD599 pKa = 3.85GVFLYY604 pKa = 10.46GHH606 pKa = 6.49GVSLIRR612 pKa = 11.84EE613 pKa = 4.39PNSSGPTFTAPSRR626 pKa = 11.84VTGWFRR632 pKa = 11.84QKK634 pKa = 9.32STYY637 pKa = 10.23YY638 pKa = 10.05VANNGSGVLHH648 pKa = 7.0LYY650 pKa = 10.68ANNEE654 pKa = 4.09GYY656 pKa = 10.4QYY658 pKa = 11.13GPTGIPTSAPIIANGINFGPASNYY682 pKa = 8.76PWLNNASLSVSVDD695 pKa = 3.22QYY697 pKa = 11.44VDD699 pKa = 3.57NIVINVNHH707 pKa = 7.26CDD709 pKa = 3.62FDD711 pKa = 5.97GDD713 pKa = 4.17GVQNSFDD720 pKa = 4.54LDD722 pKa = 3.86SDD724 pKa = 3.87NDD726 pKa = 3.96GCSDD730 pKa = 3.79SNEE733 pKa = 4.29YY734 pKa = 10.33YY735 pKa = 11.34NNATSAASGQQFGQTGGAVAPVNANGTVNLPAATYY770 pKa = 8.39TGSYY774 pKa = 11.06ANATSVGTSSSIATQPVNRR793 pKa = 11.84IINAGTNTTFSVAAAGGSGVTQYY816 pKa = 9.94QWQVDD821 pKa = 4.01SGSGFTDD828 pKa = 3.47ISNGAVYY835 pKa = 11.1NNATTNTLTLTSVPATFNGYY855 pKa = 7.57VYY857 pKa = 10.43RR858 pKa = 11.84VIIRR862 pKa = 11.84EE863 pKa = 4.31SNFVCSSVVSAEE875 pKa = 3.81RR876 pKa = 11.84TLTVFPAPVVTIADD890 pKa = 3.52ASITEE895 pKa = 4.23GGNLSFPVTLSNPSATDD912 pKa = 3.01ITVTLGFTNVTTANGDD928 pKa = 3.71YY929 pKa = 7.76TTVPVTVTFLAGATTATATVPTTADD954 pKa = 3.55TIDD957 pKa = 3.89EE958 pKa = 4.37LDD960 pKa = 3.4EE961 pKa = 4.48TFTVAITSTTGTVGSTTDD979 pKa = 3.24TATGTINDD987 pKa = 4.83DD988 pKa = 3.86DD989 pKa = 4.29NAPTVASISPSNAA1002 pKa = 2.79
Molecular weight: 103.07 kDa
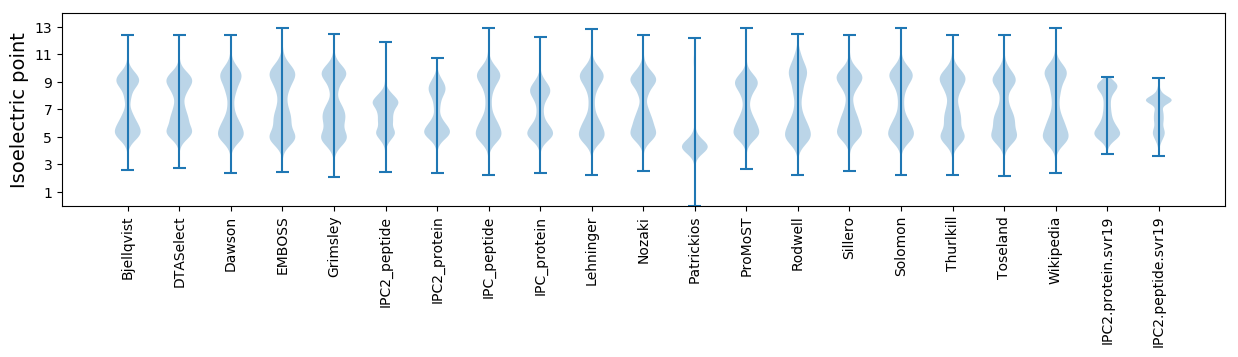
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C9ZPF7|A0A2C9ZPF7_9FLAO Alkyl hydroperoxide reductase OS=Flavobacterium sp. FPG59 OX=1929267 GN=FPG59_11905 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1053902 |
25 |
2925 |
332.1 |
37.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.61 ± 0.052 | 0.788 ± 0.015 |
5.221 ± 0.028 | 6.328 ± 0.046 |
5.416 ± 0.037 | 6.137 ± 0.042 |
1.674 ± 0.021 | 8.126 ± 0.041 |
8.013 ± 0.048 | 9.293 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.202 ± 0.024 | 6.356 ± 0.052 |
3.32 ± 0.025 | 3.635 ± 0.026 |
3.144 ± 0.031 | 6.468 ± 0.038 |
6.055 ± 0.053 | 6.323 ± 0.035 |
0.972 ± 0.015 | 3.918 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
