
Thermus phage phiOH3
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Paulinoviridae; Thomixvirus; Thermus virus OH3
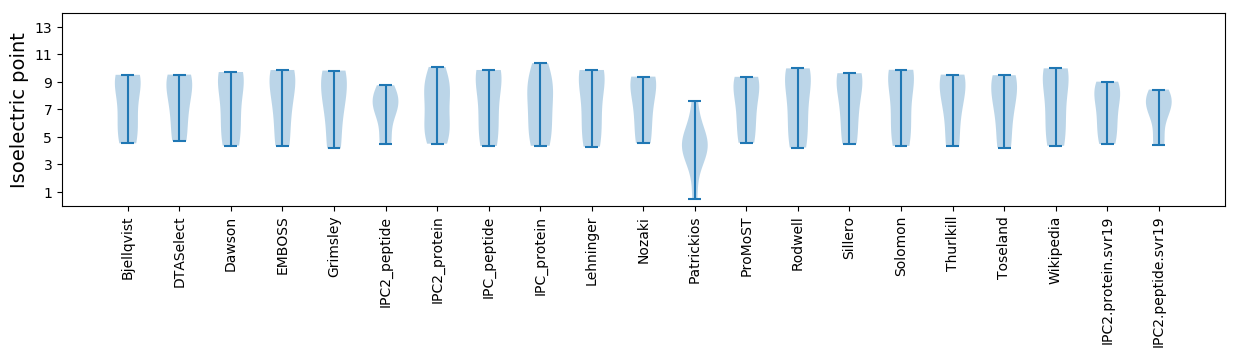
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
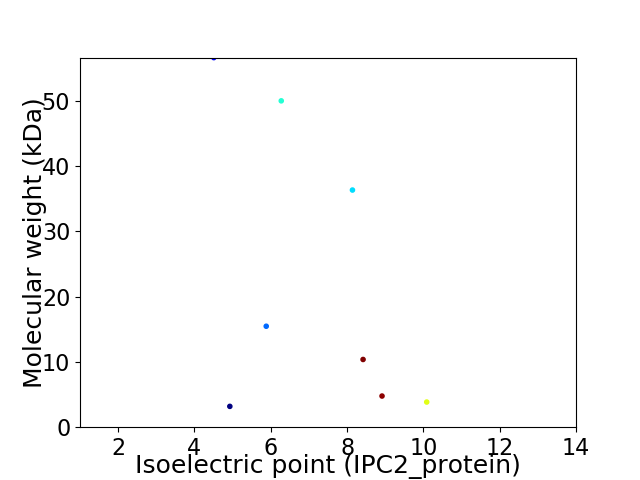
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M5N6V9|A0A0M5N6V9_9VIRU Minor coat protein OS=Thermus phage phiOH3 OX=1968339 GN=VII PE=4 SV=1
MM1 pKa = 7.43SRR3 pKa = 11.84LALPILLVLLSLGWGGPALAQPQLYY28 pKa = 9.88RR29 pKa = 11.84DD30 pKa = 4.78AIRR33 pKa = 11.84SAVQDD38 pKa = 3.45MVVAEE43 pKa = 4.54VIRR46 pKa = 11.84WPDD49 pKa = 3.08SAVRR53 pKa = 11.84KK54 pKa = 9.25SATVGRR60 pKa = 11.84YY61 pKa = 7.17VRR63 pKa = 11.84WGGAIVLGSALVYY76 pKa = 9.11TALDD80 pKa = 3.25YY81 pKa = 10.78FYY83 pKa = 11.29NVLKK87 pKa = 10.73AQTGTSLDD95 pKa = 2.93RR96 pKa = 11.84WYY98 pKa = 10.45FWSNPYY104 pKa = 10.18VEE106 pKa = 4.75VGRR109 pKa = 11.84CIDD112 pKa = 3.37RR113 pKa = 11.84GSYY116 pKa = 10.46RR117 pKa = 11.84DD118 pKa = 4.79FIAVAYY124 pKa = 10.47VNGRR128 pKa = 11.84QVNFYY133 pKa = 9.34STSSYY138 pKa = 10.65GKK140 pKa = 9.84CDD142 pKa = 3.54VPSLQSFTEE151 pKa = 3.85HH152 pKa = 6.53WYY154 pKa = 9.51NWHH157 pKa = 6.52LVRR160 pKa = 11.84NYY162 pKa = 10.88SEE164 pKa = 4.26LQGVPTQYY172 pKa = 10.77VSPPPGTCPSGWTCIAVAPQLDD194 pKa = 4.49RR195 pKa = 11.84PPLPDD200 pKa = 3.39WLQSHH205 pKa = 7.6PDD207 pKa = 3.16AADD210 pKa = 3.53GVKK213 pKa = 10.1QAVTTYY219 pKa = 11.07LDD221 pKa = 3.8RR222 pKa = 11.84NPIGSPYY229 pKa = 10.58APYY232 pKa = 10.19PGVQLEE238 pKa = 4.97PIPNPNQWTDD248 pKa = 3.29NPFTRR253 pKa = 11.84PDD255 pKa = 3.46IDD257 pKa = 3.52TDD259 pKa = 3.77GDD261 pKa = 3.69GWPDD265 pKa = 3.31PVEE268 pKa = 3.51WRR270 pKa = 11.84EE271 pKa = 3.72ANRR274 pKa = 11.84RR275 pKa = 11.84GVPWPDD281 pKa = 5.02LINNPQAYY289 pKa = 7.88PDD291 pKa = 4.32PNGDD295 pKa = 3.86PDD297 pKa = 4.24GDD299 pKa = 4.04GYY301 pKa = 9.92TNLEE305 pKa = 4.17EE306 pKa = 4.4VQQGTDD312 pKa = 3.27PYY314 pKa = 11.06DD315 pKa = 3.7PASYY319 pKa = 9.78PVRR322 pKa = 11.84RR323 pKa = 11.84SPTSPWVDD331 pKa = 3.05TDD333 pKa = 3.34GDD335 pKa = 4.29GYY337 pKa = 11.18SDD339 pKa = 3.59EE340 pKa = 4.36EE341 pKa = 4.49EE342 pKa = 3.98IRR344 pKa = 11.84KK345 pKa = 7.8GTNPNDD351 pKa = 3.54PASYY355 pKa = 10.12PDD357 pKa = 4.25TPPEE361 pKa = 3.93QQPEE365 pKa = 4.26EE366 pKa = 4.51NPDD369 pKa = 3.22EE370 pKa = 4.5PQWPGGPPSGRR381 pKa = 11.84IDD383 pKa = 3.4PVQLPEE389 pKa = 3.98VEE391 pKa = 4.04QVEE394 pKa = 4.5RR395 pKa = 11.84EE396 pKa = 4.33KK397 pKa = 11.31LPEE400 pKa = 3.83WDD402 pKa = 3.96KK403 pKa = 11.71LNPLAEE409 pKa = 4.1AWRR412 pKa = 11.84QQVVDD417 pKa = 3.9RR418 pKa = 11.84VSQKK422 pKa = 10.05FAEE425 pKa = 4.35VQNILKK431 pKa = 10.46DD432 pKa = 3.55RR433 pKa = 11.84FPFGIIAAIRR443 pKa = 11.84QRR445 pKa = 11.84VSFADD450 pKa = 3.68AQCAIQLSLPPLGTLAVDD468 pKa = 3.84ICSTPVWQLATSFRR482 pKa = 11.84PVLAGLVWVAFGFAVIRR499 pKa = 11.84RR500 pKa = 11.84ALDD503 pKa = 3.3VQKK506 pKa = 11.3
MM1 pKa = 7.43SRR3 pKa = 11.84LALPILLVLLSLGWGGPALAQPQLYY28 pKa = 9.88RR29 pKa = 11.84DD30 pKa = 4.78AIRR33 pKa = 11.84SAVQDD38 pKa = 3.45MVVAEE43 pKa = 4.54VIRR46 pKa = 11.84WPDD49 pKa = 3.08SAVRR53 pKa = 11.84KK54 pKa = 9.25SATVGRR60 pKa = 11.84YY61 pKa = 7.17VRR63 pKa = 11.84WGGAIVLGSALVYY76 pKa = 9.11TALDD80 pKa = 3.25YY81 pKa = 10.78FYY83 pKa = 11.29NVLKK87 pKa = 10.73AQTGTSLDD95 pKa = 2.93RR96 pKa = 11.84WYY98 pKa = 10.45FWSNPYY104 pKa = 10.18VEE106 pKa = 4.75VGRR109 pKa = 11.84CIDD112 pKa = 3.37RR113 pKa = 11.84GSYY116 pKa = 10.46RR117 pKa = 11.84DD118 pKa = 4.79FIAVAYY124 pKa = 10.47VNGRR128 pKa = 11.84QVNFYY133 pKa = 9.34STSSYY138 pKa = 10.65GKK140 pKa = 9.84CDD142 pKa = 3.54VPSLQSFTEE151 pKa = 3.85HH152 pKa = 6.53WYY154 pKa = 9.51NWHH157 pKa = 6.52LVRR160 pKa = 11.84NYY162 pKa = 10.88SEE164 pKa = 4.26LQGVPTQYY172 pKa = 10.77VSPPPGTCPSGWTCIAVAPQLDD194 pKa = 4.49RR195 pKa = 11.84PPLPDD200 pKa = 3.39WLQSHH205 pKa = 7.6PDD207 pKa = 3.16AADD210 pKa = 3.53GVKK213 pKa = 10.1QAVTTYY219 pKa = 11.07LDD221 pKa = 3.8RR222 pKa = 11.84NPIGSPYY229 pKa = 10.58APYY232 pKa = 10.19PGVQLEE238 pKa = 4.97PIPNPNQWTDD248 pKa = 3.29NPFTRR253 pKa = 11.84PDD255 pKa = 3.46IDD257 pKa = 3.52TDD259 pKa = 3.77GDD261 pKa = 3.69GWPDD265 pKa = 3.31PVEE268 pKa = 3.51WRR270 pKa = 11.84EE271 pKa = 3.72ANRR274 pKa = 11.84RR275 pKa = 11.84GVPWPDD281 pKa = 5.02LINNPQAYY289 pKa = 7.88PDD291 pKa = 4.32PNGDD295 pKa = 3.86PDD297 pKa = 4.24GDD299 pKa = 4.04GYY301 pKa = 9.92TNLEE305 pKa = 4.17EE306 pKa = 4.4VQQGTDD312 pKa = 3.27PYY314 pKa = 11.06DD315 pKa = 3.7PASYY319 pKa = 9.78PVRR322 pKa = 11.84RR323 pKa = 11.84SPTSPWVDD331 pKa = 3.05TDD333 pKa = 3.34GDD335 pKa = 4.29GYY337 pKa = 11.18SDD339 pKa = 3.59EE340 pKa = 4.36EE341 pKa = 4.49EE342 pKa = 3.98IRR344 pKa = 11.84KK345 pKa = 7.8GTNPNDD351 pKa = 3.54PASYY355 pKa = 10.12PDD357 pKa = 4.25TPPEE361 pKa = 3.93QQPEE365 pKa = 4.26EE366 pKa = 4.51NPDD369 pKa = 3.22EE370 pKa = 4.5PQWPGGPPSGRR381 pKa = 11.84IDD383 pKa = 3.4PVQLPEE389 pKa = 3.98VEE391 pKa = 4.04QVEE394 pKa = 4.5RR395 pKa = 11.84EE396 pKa = 4.33KK397 pKa = 11.31LPEE400 pKa = 3.83WDD402 pKa = 3.96KK403 pKa = 11.71LNPLAEE409 pKa = 4.1AWRR412 pKa = 11.84QQVVDD417 pKa = 3.9RR418 pKa = 11.84VSQKK422 pKa = 10.05FAEE425 pKa = 4.35VQNILKK431 pKa = 10.46DD432 pKa = 3.55RR433 pKa = 11.84FPFGIIAAIRR443 pKa = 11.84QRR445 pKa = 11.84VSFADD450 pKa = 3.68AQCAIQLSLPPLGTLAVDD468 pKa = 3.84ICSTPVWQLATSFRR482 pKa = 11.84PVLAGLVWVAFGFAVIRR499 pKa = 11.84RR500 pKa = 11.84ALDD503 pKa = 3.3VQKK506 pKa = 11.3
Molecular weight: 56.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M4VAS8|A0A0M4VAS8_9VIRU Minor coat protein OS=Thermus phage phiOH3 OX=1968339 GN=VI PE=4 SV=1
MM1 pKa = 7.48EE2 pKa = 5.55FDD4 pKa = 3.5AGAIVSTVQNYY15 pKa = 9.28ILAIAGAGVALLGLTIGLSAAWRR38 pKa = 11.84YY39 pKa = 9.02AKK41 pKa = 10.47KK42 pKa = 10.14FLKK45 pKa = 10.68GG46 pKa = 3.23
MM1 pKa = 7.48EE2 pKa = 5.55FDD4 pKa = 3.5AGAIVSTVQNYY15 pKa = 9.28ILAIAGAGVALLGLTIGLSAAWRR38 pKa = 11.84YY39 pKa = 9.02AKK41 pKa = 10.47KK42 pKa = 10.14FLKK45 pKa = 10.68GG46 pKa = 3.23
Molecular weight: 4.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1619 |
30 |
506 |
202.4 |
22.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.068 ± 1.185 | 0.556 ± 0.285 |
5.806 ± 0.911 | 5.188 ± 0.84 |
3.644 ± 0.477 | 7.844 ± 0.809 |
1.112 ± 0.272 | 3.83 ± 0.989 |
3.027 ± 0.592 | 10.377 ± 1.639 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.606 ± 0.466 | 2.347 ± 0.619 |
7.288 ± 1.56 | 3.582 ± 0.875 |
7.041 ± 1.002 | 5.683 ± 0.556 |
3.891 ± 0.53 | 9.574 ± 0.926 |
3.582 ± 0.426 | 3.953 ± 0.54 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
