
Blautia hydrogenotrophica CAG:147
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Blautia; environmental samples
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
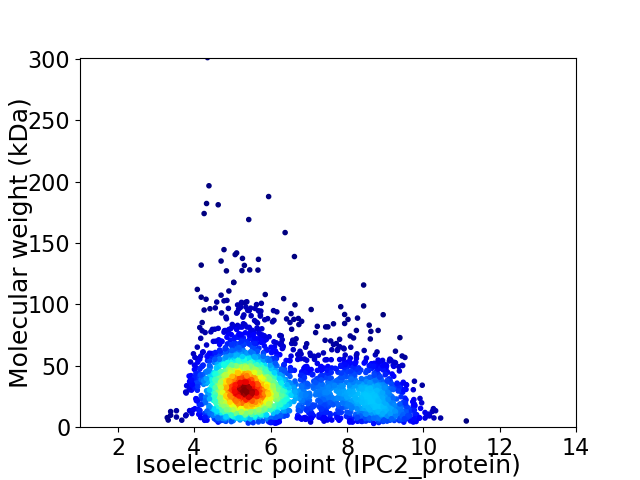
Virtual 2D-PAGE plot for 3087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5C6G8|R5C6G8_9FIRM ABC transporter domain-containing protein OS=Blautia hydrogenotrophica CAG:147 OX=1263061 GN=BN499_00718 PE=3 SV=1
MM1 pKa = 7.63KK2 pKa = 10.66SNLEE6 pKa = 3.96QEE8 pKa = 4.57LQRR11 pKa = 11.84QLRR14 pKa = 11.84QEE16 pKa = 3.94IEE18 pKa = 3.9IPEE21 pKa = 4.64CIQKK25 pKa = 10.33KK26 pKa = 10.07SKK28 pKa = 9.68QAYY31 pKa = 9.45LKK33 pKa = 10.18IKK35 pKa = 9.96SGQIRR40 pKa = 11.84QEE42 pKa = 4.16EE43 pKa = 4.56VLKK46 pKa = 10.24TPLGWMKK53 pKa = 9.54KK54 pKa = 6.47TAIALGSVAAALLLCMVLFLTNPVMAQDD82 pKa = 4.26FPVIGNLFSLLQDD95 pKa = 4.01KK96 pKa = 10.86ISFFGNFEE104 pKa = 4.67DD105 pKa = 4.66YY106 pKa = 8.43ATPLVPEE113 pKa = 4.68EE114 pKa = 4.28DD115 pKa = 3.46TSFSKK120 pKa = 10.32DD121 pKa = 3.13TDD123 pKa = 3.45SSGIFSKK130 pKa = 9.96TEE132 pKa = 4.05DD133 pKa = 3.83GLTITLSEE141 pKa = 4.69VYY143 pKa = 10.8ANTQAIYY150 pKa = 8.95LTVLMEE156 pKa = 4.27SEE158 pKa = 4.24EE159 pKa = 4.4AFPSTLIDD167 pKa = 3.58QQDD170 pKa = 3.6QPVLSLIASEE180 pKa = 5.27NYY182 pKa = 10.17DD183 pKa = 3.74FNPTPEE189 pKa = 4.46EE190 pKa = 4.19YY191 pKa = 10.93NNPIYY196 pKa = 10.28FNPEE200 pKa = 3.44GQFLDD205 pKa = 4.62DD206 pKa = 3.73YY207 pKa = 10.42TYY209 pKa = 11.53SCILRR214 pKa = 11.84LDD216 pKa = 3.64LRR218 pKa = 11.84EE219 pKa = 4.3ASLDD223 pKa = 3.34RR224 pKa = 11.84QAYY227 pKa = 8.01NEE229 pKa = 3.97AMIQAEE235 pKa = 4.27AAAQEE240 pKa = 3.95TDD242 pKa = 4.19AVINYY247 pKa = 9.11QNLQSFVEE255 pKa = 4.31IPEE258 pKa = 4.18QIHH261 pKa = 7.19LDD263 pKa = 3.7FQIQQVIGTLEE274 pKa = 4.01DD275 pKa = 3.16AQYY278 pKa = 10.21WDD280 pKa = 2.86SGYY283 pKa = 9.16TEE285 pKa = 4.69EE286 pKa = 5.27EE287 pKa = 4.06LTAMTDD293 pKa = 3.4EE294 pKa = 4.64EE295 pKa = 4.35FQEE298 pKa = 4.49VMKK301 pKa = 10.67QMPEE305 pKa = 4.03EE306 pKa = 4.31YY307 pKa = 9.64NQHH310 pKa = 6.0PNQYY314 pKa = 9.25EE315 pKa = 4.24NYY317 pKa = 8.58WFDD320 pKa = 4.03GPWDD324 pKa = 4.07FSLDD328 pKa = 3.61LTVDD332 pKa = 3.63SSRR335 pKa = 11.84VQTIEE340 pKa = 3.51INEE343 pKa = 3.95INEE346 pKa = 4.08NGVGISNVILTPYY359 pKa = 10.13EE360 pKa = 3.95LTVNDD365 pKa = 4.78AYY367 pKa = 10.99ADD369 pKa = 3.68GVPFYY374 pKa = 11.01DD375 pKa = 3.85YY376 pKa = 11.31FLVALDD382 pKa = 3.9ANGNKK387 pKa = 9.97LPYY390 pKa = 9.87NDD392 pKa = 4.95SNGNCNYY399 pKa = 8.66FTIQDD404 pKa = 3.71RR405 pKa = 11.84DD406 pKa = 3.24ISTVDD411 pKa = 4.7LYY413 pKa = 10.9ILDD416 pKa = 3.61YY417 pKa = 10.37TEE419 pKa = 5.77YY420 pKa = 9.57MDD422 pKa = 4.15EE423 pKa = 4.88LKK425 pKa = 11.23GEE427 pKa = 4.06EE428 pKa = 4.07NYY430 pKa = 11.13NNNEE434 pKa = 4.05NKK436 pKa = 10.1PEE438 pKa = 3.95EE439 pKa = 4.6EE440 pKa = 3.97KK441 pKa = 10.64WSTLLDD447 pKa = 3.76DD448 pKa = 3.7RR449 pKa = 11.84CFYY452 pKa = 10.13HH453 pKa = 5.69TTLNLTPVEE462 pKa = 4.17
MM1 pKa = 7.63KK2 pKa = 10.66SNLEE6 pKa = 3.96QEE8 pKa = 4.57LQRR11 pKa = 11.84QLRR14 pKa = 11.84QEE16 pKa = 3.94IEE18 pKa = 3.9IPEE21 pKa = 4.64CIQKK25 pKa = 10.33KK26 pKa = 10.07SKK28 pKa = 9.68QAYY31 pKa = 9.45LKK33 pKa = 10.18IKK35 pKa = 9.96SGQIRR40 pKa = 11.84QEE42 pKa = 4.16EE43 pKa = 4.56VLKK46 pKa = 10.24TPLGWMKK53 pKa = 9.54KK54 pKa = 6.47TAIALGSVAAALLLCMVLFLTNPVMAQDD82 pKa = 4.26FPVIGNLFSLLQDD95 pKa = 4.01KK96 pKa = 10.86ISFFGNFEE104 pKa = 4.67DD105 pKa = 4.66YY106 pKa = 8.43ATPLVPEE113 pKa = 4.68EE114 pKa = 4.28DD115 pKa = 3.46TSFSKK120 pKa = 10.32DD121 pKa = 3.13TDD123 pKa = 3.45SSGIFSKK130 pKa = 9.96TEE132 pKa = 4.05DD133 pKa = 3.83GLTITLSEE141 pKa = 4.69VYY143 pKa = 10.8ANTQAIYY150 pKa = 8.95LTVLMEE156 pKa = 4.27SEE158 pKa = 4.24EE159 pKa = 4.4AFPSTLIDD167 pKa = 3.58QQDD170 pKa = 3.6QPVLSLIASEE180 pKa = 5.27NYY182 pKa = 10.17DD183 pKa = 3.74FNPTPEE189 pKa = 4.46EE190 pKa = 4.19YY191 pKa = 10.93NNPIYY196 pKa = 10.28FNPEE200 pKa = 3.44GQFLDD205 pKa = 4.62DD206 pKa = 3.73YY207 pKa = 10.42TYY209 pKa = 11.53SCILRR214 pKa = 11.84LDD216 pKa = 3.64LRR218 pKa = 11.84EE219 pKa = 4.3ASLDD223 pKa = 3.34RR224 pKa = 11.84QAYY227 pKa = 8.01NEE229 pKa = 3.97AMIQAEE235 pKa = 4.27AAAQEE240 pKa = 3.95TDD242 pKa = 4.19AVINYY247 pKa = 9.11QNLQSFVEE255 pKa = 4.31IPEE258 pKa = 4.18QIHH261 pKa = 7.19LDD263 pKa = 3.7FQIQQVIGTLEE274 pKa = 4.01DD275 pKa = 3.16AQYY278 pKa = 10.21WDD280 pKa = 2.86SGYY283 pKa = 9.16TEE285 pKa = 4.69EE286 pKa = 5.27EE287 pKa = 4.06LTAMTDD293 pKa = 3.4EE294 pKa = 4.64EE295 pKa = 4.35FQEE298 pKa = 4.49VMKK301 pKa = 10.67QMPEE305 pKa = 4.03EE306 pKa = 4.31YY307 pKa = 9.64NQHH310 pKa = 6.0PNQYY314 pKa = 9.25EE315 pKa = 4.24NYY317 pKa = 8.58WFDD320 pKa = 4.03GPWDD324 pKa = 4.07FSLDD328 pKa = 3.61LTVDD332 pKa = 3.63SSRR335 pKa = 11.84VQTIEE340 pKa = 3.51INEE343 pKa = 3.95INEE346 pKa = 4.08NGVGISNVILTPYY359 pKa = 10.13EE360 pKa = 3.95LTVNDD365 pKa = 4.78AYY367 pKa = 10.99ADD369 pKa = 3.68GVPFYY374 pKa = 11.01DD375 pKa = 3.85YY376 pKa = 11.31FLVALDD382 pKa = 3.9ANGNKK387 pKa = 9.97LPYY390 pKa = 9.87NDD392 pKa = 4.95SNGNCNYY399 pKa = 8.66FTIQDD404 pKa = 3.71RR405 pKa = 11.84DD406 pKa = 3.24ISTVDD411 pKa = 4.7LYY413 pKa = 10.9ILDD416 pKa = 3.61YY417 pKa = 10.37TEE419 pKa = 5.77YY420 pKa = 9.57MDD422 pKa = 4.15EE423 pKa = 4.88LKK425 pKa = 11.23GEE427 pKa = 4.06EE428 pKa = 4.07NYY430 pKa = 11.13NNNEE434 pKa = 4.05NKK436 pKa = 10.1PEE438 pKa = 3.95EE439 pKa = 4.6EE440 pKa = 3.97KK441 pKa = 10.64WSTLLDD447 pKa = 3.76DD448 pKa = 3.7RR449 pKa = 11.84CFYY452 pKa = 10.13HH453 pKa = 5.69TTLNLTPVEE462 pKa = 4.17
Molecular weight: 53.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5C5M1|R5C5M1_9FIRM Uncharacterized protein OS=Blautia hydrogenotrophica CAG:147 OX=1263061 GN=BN499_02185 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
947112 |
29 |
2797 |
306.8 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.315 ± 0.046 | 1.642 ± 0.019 |
5.117 ± 0.031 | 7.908 ± 0.05 |
4.018 ± 0.031 | 7.467 ± 0.043 |
1.717 ± 0.023 | 6.974 ± 0.038 |
6.578 ± 0.035 | 9.373 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.088 ± 0.023 | 3.907 ± 0.025 |
3.391 ± 0.024 | 3.645 ± 0.03 |
4.741 ± 0.037 | 5.742 ± 0.03 |
5.173 ± 0.032 | 7.259 ± 0.036 |
0.933 ± 0.016 | 3.999 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
