
Mariniphaga anaerophila
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Mariniphaga
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
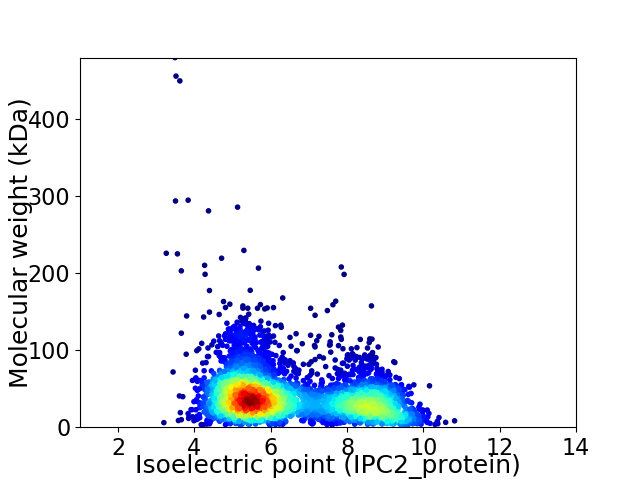
Virtual 2D-PAGE plot for 3974 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5ETJ0|A0A1M5ETJ0_9BACT Uncharacterized protein OS=Mariniphaga anaerophila OX=1484053 GN=SAMN05444274_11021 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.24KK3 pKa = 10.3FYY5 pKa = 10.51FFSFFALLFFIGSGLTFAAEE25 pKa = 3.69IGDD28 pKa = 4.0NIKK31 pKa = 10.39IGSNQQSAKK40 pKa = 10.12FPQLTYY46 pKa = 10.63CSEE49 pKa = 3.78QDD51 pKa = 3.59KK52 pKa = 11.26ILMVWNGNDD61 pKa = 3.28SGTDD65 pKa = 3.12QIFFKK70 pKa = 10.71FLTHH74 pKa = 6.8SEE76 pKa = 4.32VSQNPSAPGMQLSSGSSPWNRR97 pKa = 11.84YY98 pKa = 6.9SEE100 pKa = 4.47RR101 pKa = 11.84PRR103 pKa = 11.84AAYY106 pKa = 9.71DD107 pKa = 3.44AFSATAAVVWQEE119 pKa = 3.99SNAVGKK125 pKa = 10.35DD126 pKa = 3.55SVVMAILDD134 pKa = 3.74VNTEE138 pKa = 4.03SKK140 pKa = 9.63LTEE143 pKa = 3.95VVVAANSSWNMTATVASDD161 pKa = 3.5NNGSFLVAYY170 pKa = 9.75YY171 pKa = 10.79DD172 pKa = 3.71SASTNITAKK181 pKa = 10.83LFDD184 pKa = 4.44ANGAQSGSDD193 pKa = 3.6VILGTVAGGYY203 pKa = 7.24MFPYY207 pKa = 10.23SIDD210 pKa = 2.95IAYY213 pKa = 10.52NSEE216 pKa = 3.91NEE218 pKa = 3.99IFLVTWADD226 pKa = 3.58YY227 pKa = 11.18NSNLFSRR234 pKa = 11.84TLNSDD239 pKa = 3.4GTAGEE244 pKa = 4.24LNVYY248 pKa = 10.7NSIQGVANPSVAYY261 pKa = 10.19NDD263 pKa = 3.67SLNIFLIAYY272 pKa = 9.72DD273 pKa = 4.01DD274 pKa = 3.85FAGNVSAQLTDD285 pKa = 3.12SLGILSGSLFTFGSSSALEE304 pKa = 4.28AEE306 pKa = 4.28PDD308 pKa = 3.27VAYY311 pKa = 10.46NAKK314 pKa = 10.45NNGFAVTWSDD324 pKa = 3.61YY325 pKa = 11.32LGGSGVWVQEE335 pKa = 4.1ILVSSCTVLVDD346 pKa = 3.68SVKK349 pKa = 9.75MDD351 pKa = 4.41KK352 pKa = 10.59ISATTNSSSVAYY364 pKa = 10.15DD365 pKa = 3.6AVNDD369 pKa = 4.28SYY371 pKa = 10.94WVAWYY376 pKa = 10.5GEE378 pKa = 4.23SGGAKK383 pKa = 10.21DD384 pKa = 3.98EE385 pKa = 5.08LFLQRR390 pKa = 11.84YY391 pKa = 7.72RR392 pKa = 11.84SEE394 pKa = 4.47NISIKK399 pKa = 10.71ANCKK403 pKa = 10.47DD404 pKa = 2.77ITVYY408 pKa = 10.88LDD410 pKa = 3.31EE411 pKa = 5.31NGEE414 pKa = 4.38YY415 pKa = 10.47VLSAEE420 pKa = 4.65DD421 pKa = 3.43VDD423 pKa = 4.36NGSFLLCGHH432 pKa = 5.89VQMDD436 pKa = 3.46IDD438 pKa = 3.64VDD440 pKa = 3.89EE441 pKa = 5.33ALTCAVTQPVEE452 pKa = 4.1VRR454 pKa = 11.84LTASDD459 pKa = 3.81GDD461 pKa = 4.05EE462 pKa = 3.95QTDD465 pKa = 3.97YY466 pKa = 11.44CVATVTVLDD475 pKa = 4.08TVPPVPVCEE484 pKa = 4.18NKK486 pKa = 10.33TIFLDD491 pKa = 4.12ANGEE495 pKa = 4.16AYY497 pKa = 7.59LTPEE501 pKa = 5.32DD502 pKa = 4.12IDD504 pKa = 4.56GGSTDD509 pKa = 3.01NCGIQAIWLDD519 pKa = 3.32KK520 pKa = 10.6TDD522 pKa = 5.54FYY524 pKa = 11.25TADD527 pKa = 3.52LGEE530 pKa = 4.28NTVLLTVKK538 pKa = 10.43DD539 pKa = 3.54NAGIEE544 pKa = 4.24RR545 pKa = 11.84SCEE548 pKa = 3.94AIVTVSEE555 pKa = 5.37FIDD558 pKa = 3.71DD559 pKa = 3.96TPPSVRR565 pKa = 11.84TTPLTIYY572 pKa = 10.66LDD574 pKa = 3.83EE575 pKa = 4.91TGTASIEE582 pKa = 4.05PSDD585 pKa = 3.79IDD587 pKa = 4.31NGSFDD592 pKa = 3.98DD593 pKa = 5.7GEE595 pKa = 4.37IEE597 pKa = 4.92SLSLDD602 pKa = 3.44KK603 pKa = 11.31SVFTCDD609 pKa = 3.39DD610 pKa = 3.58ANRR613 pKa = 11.84TDD615 pKa = 3.4TMFALAPEE623 pKa = 4.62ASTGSVARR631 pKa = 11.84FVGSQGKK638 pKa = 8.42DD639 pKa = 2.56WKK641 pKa = 10.19YY642 pKa = 11.2DD643 pKa = 3.31GVAGSTGLGFNYY655 pKa = 10.0GFEE658 pKa = 4.68RR659 pKa = 11.84NLSDD663 pKa = 3.16GQYY666 pKa = 10.32YY667 pKa = 10.77LIGGASEE674 pKa = 4.71DD675 pKa = 3.53EE676 pKa = 4.28RR677 pKa = 11.84NLYY680 pKa = 10.4RR681 pKa = 11.84LDD683 pKa = 4.12AEE685 pKa = 4.47TFSAEE690 pKa = 4.4FITEE694 pKa = 4.33LMSSDD699 pKa = 3.57GDD701 pKa = 3.93EE702 pKa = 4.49EE703 pKa = 4.39PLDD706 pKa = 3.79IAIDD710 pKa = 3.55NNGYY714 pKa = 9.85FFIVYY719 pKa = 10.34KK720 pKa = 10.84NGTIDD725 pKa = 4.16KK726 pKa = 9.39LDD728 pKa = 3.99PVTWTAVEE736 pKa = 4.08HH737 pKa = 6.31ANIGAGLGEE746 pKa = 4.33VSLTYY751 pKa = 10.83DD752 pKa = 3.49NEE754 pKa = 4.07NEE756 pKa = 3.92RR757 pKa = 11.84LLFVHH762 pKa = 6.93FLQNEE767 pKa = 4.19AVNAPNVLSEE777 pKa = 3.92ISSNGTVTEE786 pKa = 4.28LFSFFTPGNDD796 pKa = 3.63SSCCALGIEE805 pKa = 4.47YY806 pKa = 10.2AGNGFCFASSAFNCDD821 pKa = 2.95ALFEE825 pKa = 5.19IDD827 pKa = 5.34LNNEE831 pKa = 4.12SVNLIAQPNGSSFSGLKK848 pKa = 10.34DD849 pKa = 4.43LIYY852 pKa = 10.47IPANMVTLIVTDD864 pKa = 3.84IYY866 pKa = 11.48GNSSEE871 pKa = 4.67GKK873 pKa = 9.08VKK875 pKa = 11.01VEE877 pKa = 3.78VVDD880 pKa = 4.5NIEE883 pKa = 4.02PALVLNQVQVYY894 pKa = 9.18LGPDD898 pKa = 3.24GTYY901 pKa = 10.48KK902 pKa = 9.37ITEE905 pKa = 3.86QDD907 pKa = 3.32IEE909 pKa = 4.36AMIAGTDD916 pKa = 3.99DD917 pKa = 3.65NCSSSGNLEE926 pKa = 3.8FEE928 pKa = 4.67VYY930 pKa = 10.2PKK932 pKa = 10.46IFDD935 pKa = 3.63CTMDD939 pKa = 4.1EE940 pKa = 4.2IVHH943 pKa = 5.28VRR945 pKa = 11.84VTAADD950 pKa = 3.14ASGNYY955 pKa = 7.26TRR957 pKa = 11.84AWTTVLVFDD966 pKa = 4.05TLSPVFEE973 pKa = 4.25PVADD977 pKa = 4.45FEE979 pKa = 5.13TTVSPGTSEE988 pKa = 4.13TPIEE992 pKa = 4.27YY993 pKa = 10.04PEE995 pKa = 4.1VLATDD1000 pKa = 3.59NCSVEE1005 pKa = 4.13VNLIGGLGPNGLFPVGVTTEE1025 pKa = 3.47KK1026 pKa = 10.17WVAVDD1031 pKa = 4.97DD1032 pKa = 4.67RR1033 pKa = 11.84GNSDD1037 pKa = 3.04TLMFNVTVVALNANPTINPISDD1059 pKa = 3.74VEE1061 pKa = 4.41VSEE1064 pKa = 4.37STSPITVEE1072 pKa = 4.12LAGISYY1078 pKa = 10.98GNDD1081 pKa = 2.72LVEE1084 pKa = 4.08QTVAVTAKK1092 pKa = 10.33ADD1094 pKa = 3.66NVDD1097 pKa = 3.83LVEE1100 pKa = 4.94SIFVNYY1106 pKa = 10.02EE1107 pKa = 3.8SGSTGSLEE1115 pKa = 3.89IEE1117 pKa = 4.18IASEE1121 pKa = 3.95RR1122 pKa = 11.84SGVAIITVNVIDD1134 pKa = 4.05SEE1136 pKa = 4.31GGEE1139 pKa = 4.0VSEE1142 pKa = 4.42TFRR1145 pKa = 11.84LTVIPVNKK1153 pKa = 9.68PPFLVYY1159 pKa = 10.07PISDD1163 pKa = 3.01QWVNASYY1170 pKa = 10.73VLKK1173 pKa = 11.0VPVSPVLGEE1182 pKa = 4.72LFDD1185 pKa = 5.46DD1186 pKa = 4.65PEE1188 pKa = 6.41GDD1190 pKa = 3.78ALTISAMLADD1200 pKa = 4.23GTVLPGWAEE1209 pKa = 3.93LVNDD1213 pKa = 3.92SLVFNPMIEE1222 pKa = 4.18NVGCIDD1228 pKa = 3.93VVVKK1232 pKa = 9.99ATDD1235 pKa = 3.35IEE1237 pKa = 4.78GGSATDD1243 pKa = 3.45TFAVCVEE1250 pKa = 4.74GYY1252 pKa = 9.27PVNADD1257 pKa = 3.88AISAGGLSVNLYY1269 pKa = 9.44PNPAVDD1275 pKa = 4.6KK1276 pKa = 10.72VNISIKK1282 pKa = 10.34SSSFEE1287 pKa = 3.94PTVVTVYY1294 pKa = 10.33TITGEE1299 pKa = 4.11LVLQGKK1305 pKa = 9.44FDD1307 pKa = 4.07AGQLISLDD1315 pKa = 3.66MAEE1318 pKa = 4.79HH1319 pKa = 5.78VSGVYY1324 pKa = 8.9FVKK1327 pKa = 9.97IDD1329 pKa = 3.42QNNRR1333 pKa = 11.84TVVKK1337 pKa = 10.59KK1338 pKa = 10.62LVLKK1342 pKa = 10.93GKK1344 pKa = 10.05
MM1 pKa = 7.41KK2 pKa = 10.24KK3 pKa = 10.3FYY5 pKa = 10.51FFSFFALLFFIGSGLTFAAEE25 pKa = 3.69IGDD28 pKa = 4.0NIKK31 pKa = 10.39IGSNQQSAKK40 pKa = 10.12FPQLTYY46 pKa = 10.63CSEE49 pKa = 3.78QDD51 pKa = 3.59KK52 pKa = 11.26ILMVWNGNDD61 pKa = 3.28SGTDD65 pKa = 3.12QIFFKK70 pKa = 10.71FLTHH74 pKa = 6.8SEE76 pKa = 4.32VSQNPSAPGMQLSSGSSPWNRR97 pKa = 11.84YY98 pKa = 6.9SEE100 pKa = 4.47RR101 pKa = 11.84PRR103 pKa = 11.84AAYY106 pKa = 9.71DD107 pKa = 3.44AFSATAAVVWQEE119 pKa = 3.99SNAVGKK125 pKa = 10.35DD126 pKa = 3.55SVVMAILDD134 pKa = 3.74VNTEE138 pKa = 4.03SKK140 pKa = 9.63LTEE143 pKa = 3.95VVVAANSSWNMTATVASDD161 pKa = 3.5NNGSFLVAYY170 pKa = 9.75YY171 pKa = 10.79DD172 pKa = 3.71SASTNITAKK181 pKa = 10.83LFDD184 pKa = 4.44ANGAQSGSDD193 pKa = 3.6VILGTVAGGYY203 pKa = 7.24MFPYY207 pKa = 10.23SIDD210 pKa = 2.95IAYY213 pKa = 10.52NSEE216 pKa = 3.91NEE218 pKa = 3.99IFLVTWADD226 pKa = 3.58YY227 pKa = 11.18NSNLFSRR234 pKa = 11.84TLNSDD239 pKa = 3.4GTAGEE244 pKa = 4.24LNVYY248 pKa = 10.7NSIQGVANPSVAYY261 pKa = 10.19NDD263 pKa = 3.67SLNIFLIAYY272 pKa = 9.72DD273 pKa = 4.01DD274 pKa = 3.85FAGNVSAQLTDD285 pKa = 3.12SLGILSGSLFTFGSSSALEE304 pKa = 4.28AEE306 pKa = 4.28PDD308 pKa = 3.27VAYY311 pKa = 10.46NAKK314 pKa = 10.45NNGFAVTWSDD324 pKa = 3.61YY325 pKa = 11.32LGGSGVWVQEE335 pKa = 4.1ILVSSCTVLVDD346 pKa = 3.68SVKK349 pKa = 9.75MDD351 pKa = 4.41KK352 pKa = 10.59ISATTNSSSVAYY364 pKa = 10.15DD365 pKa = 3.6AVNDD369 pKa = 4.28SYY371 pKa = 10.94WVAWYY376 pKa = 10.5GEE378 pKa = 4.23SGGAKK383 pKa = 10.21DD384 pKa = 3.98EE385 pKa = 5.08LFLQRR390 pKa = 11.84YY391 pKa = 7.72RR392 pKa = 11.84SEE394 pKa = 4.47NISIKK399 pKa = 10.71ANCKK403 pKa = 10.47DD404 pKa = 2.77ITVYY408 pKa = 10.88LDD410 pKa = 3.31EE411 pKa = 5.31NGEE414 pKa = 4.38YY415 pKa = 10.47VLSAEE420 pKa = 4.65DD421 pKa = 3.43VDD423 pKa = 4.36NGSFLLCGHH432 pKa = 5.89VQMDD436 pKa = 3.46IDD438 pKa = 3.64VDD440 pKa = 3.89EE441 pKa = 5.33ALTCAVTQPVEE452 pKa = 4.1VRR454 pKa = 11.84LTASDD459 pKa = 3.81GDD461 pKa = 4.05EE462 pKa = 3.95QTDD465 pKa = 3.97YY466 pKa = 11.44CVATVTVLDD475 pKa = 4.08TVPPVPVCEE484 pKa = 4.18NKK486 pKa = 10.33TIFLDD491 pKa = 4.12ANGEE495 pKa = 4.16AYY497 pKa = 7.59LTPEE501 pKa = 5.32DD502 pKa = 4.12IDD504 pKa = 4.56GGSTDD509 pKa = 3.01NCGIQAIWLDD519 pKa = 3.32KK520 pKa = 10.6TDD522 pKa = 5.54FYY524 pKa = 11.25TADD527 pKa = 3.52LGEE530 pKa = 4.28NTVLLTVKK538 pKa = 10.43DD539 pKa = 3.54NAGIEE544 pKa = 4.24RR545 pKa = 11.84SCEE548 pKa = 3.94AIVTVSEE555 pKa = 5.37FIDD558 pKa = 3.71DD559 pKa = 3.96TPPSVRR565 pKa = 11.84TTPLTIYY572 pKa = 10.66LDD574 pKa = 3.83EE575 pKa = 4.91TGTASIEE582 pKa = 4.05PSDD585 pKa = 3.79IDD587 pKa = 4.31NGSFDD592 pKa = 3.98DD593 pKa = 5.7GEE595 pKa = 4.37IEE597 pKa = 4.92SLSLDD602 pKa = 3.44KK603 pKa = 11.31SVFTCDD609 pKa = 3.39DD610 pKa = 3.58ANRR613 pKa = 11.84TDD615 pKa = 3.4TMFALAPEE623 pKa = 4.62ASTGSVARR631 pKa = 11.84FVGSQGKK638 pKa = 8.42DD639 pKa = 2.56WKK641 pKa = 10.19YY642 pKa = 11.2DD643 pKa = 3.31GVAGSTGLGFNYY655 pKa = 10.0GFEE658 pKa = 4.68RR659 pKa = 11.84NLSDD663 pKa = 3.16GQYY666 pKa = 10.32YY667 pKa = 10.77LIGGASEE674 pKa = 4.71DD675 pKa = 3.53EE676 pKa = 4.28RR677 pKa = 11.84NLYY680 pKa = 10.4RR681 pKa = 11.84LDD683 pKa = 4.12AEE685 pKa = 4.47TFSAEE690 pKa = 4.4FITEE694 pKa = 4.33LMSSDD699 pKa = 3.57GDD701 pKa = 3.93EE702 pKa = 4.49EE703 pKa = 4.39PLDD706 pKa = 3.79IAIDD710 pKa = 3.55NNGYY714 pKa = 9.85FFIVYY719 pKa = 10.34KK720 pKa = 10.84NGTIDD725 pKa = 4.16KK726 pKa = 9.39LDD728 pKa = 3.99PVTWTAVEE736 pKa = 4.08HH737 pKa = 6.31ANIGAGLGEE746 pKa = 4.33VSLTYY751 pKa = 10.83DD752 pKa = 3.49NEE754 pKa = 4.07NEE756 pKa = 3.92RR757 pKa = 11.84LLFVHH762 pKa = 6.93FLQNEE767 pKa = 4.19AVNAPNVLSEE777 pKa = 3.92ISSNGTVTEE786 pKa = 4.28LFSFFTPGNDD796 pKa = 3.63SSCCALGIEE805 pKa = 4.47YY806 pKa = 10.2AGNGFCFASSAFNCDD821 pKa = 2.95ALFEE825 pKa = 5.19IDD827 pKa = 5.34LNNEE831 pKa = 4.12SVNLIAQPNGSSFSGLKK848 pKa = 10.34DD849 pKa = 4.43LIYY852 pKa = 10.47IPANMVTLIVTDD864 pKa = 3.84IYY866 pKa = 11.48GNSSEE871 pKa = 4.67GKK873 pKa = 9.08VKK875 pKa = 11.01VEE877 pKa = 3.78VVDD880 pKa = 4.5NIEE883 pKa = 4.02PALVLNQVQVYY894 pKa = 9.18LGPDD898 pKa = 3.24GTYY901 pKa = 10.48KK902 pKa = 9.37ITEE905 pKa = 3.86QDD907 pKa = 3.32IEE909 pKa = 4.36AMIAGTDD916 pKa = 3.99DD917 pKa = 3.65NCSSSGNLEE926 pKa = 3.8FEE928 pKa = 4.67VYY930 pKa = 10.2PKK932 pKa = 10.46IFDD935 pKa = 3.63CTMDD939 pKa = 4.1EE940 pKa = 4.2IVHH943 pKa = 5.28VRR945 pKa = 11.84VTAADD950 pKa = 3.14ASGNYY955 pKa = 7.26TRR957 pKa = 11.84AWTTVLVFDD966 pKa = 4.05TLSPVFEE973 pKa = 4.25PVADD977 pKa = 4.45FEE979 pKa = 5.13TTVSPGTSEE988 pKa = 4.13TPIEE992 pKa = 4.27YY993 pKa = 10.04PEE995 pKa = 4.1VLATDD1000 pKa = 3.59NCSVEE1005 pKa = 4.13VNLIGGLGPNGLFPVGVTTEE1025 pKa = 3.47KK1026 pKa = 10.17WVAVDD1031 pKa = 4.97DD1032 pKa = 4.67RR1033 pKa = 11.84GNSDD1037 pKa = 3.04TLMFNVTVVALNANPTINPISDD1059 pKa = 3.74VEE1061 pKa = 4.41VSEE1064 pKa = 4.37STSPITVEE1072 pKa = 4.12LAGISYY1078 pKa = 10.98GNDD1081 pKa = 2.72LVEE1084 pKa = 4.08QTVAVTAKK1092 pKa = 10.33ADD1094 pKa = 3.66NVDD1097 pKa = 3.83LVEE1100 pKa = 4.94SIFVNYY1106 pKa = 10.02EE1107 pKa = 3.8SGSTGSLEE1115 pKa = 3.89IEE1117 pKa = 4.18IASEE1121 pKa = 3.95RR1122 pKa = 11.84SGVAIITVNVIDD1134 pKa = 4.05SEE1136 pKa = 4.31GGEE1139 pKa = 4.0VSEE1142 pKa = 4.42TFRR1145 pKa = 11.84LTVIPVNKK1153 pKa = 9.68PPFLVYY1159 pKa = 10.07PISDD1163 pKa = 3.01QWVNASYY1170 pKa = 10.73VLKK1173 pKa = 11.0VPVSPVLGEE1182 pKa = 4.72LFDD1185 pKa = 5.46DD1186 pKa = 4.65PEE1188 pKa = 6.41GDD1190 pKa = 3.78ALTISAMLADD1200 pKa = 4.23GTVLPGWAEE1209 pKa = 3.93LVNDD1213 pKa = 3.92SLVFNPMIEE1222 pKa = 4.18NVGCIDD1228 pKa = 3.93VVVKK1232 pKa = 9.99ATDD1235 pKa = 3.35IEE1237 pKa = 4.78GGSATDD1243 pKa = 3.45TFAVCVEE1250 pKa = 4.74GYY1252 pKa = 9.27PVNADD1257 pKa = 3.88AISAGGLSVNLYY1269 pKa = 9.44PNPAVDD1275 pKa = 4.6KK1276 pKa = 10.72VNISIKK1282 pKa = 10.34SSSFEE1287 pKa = 3.94PTVVTVYY1294 pKa = 10.33TITGEE1299 pKa = 4.11LVLQGKK1305 pKa = 9.44FDD1307 pKa = 4.07AGQLISLDD1315 pKa = 3.66MAEE1318 pKa = 4.79HH1319 pKa = 5.78VSGVYY1324 pKa = 8.9FVKK1327 pKa = 9.97IDD1329 pKa = 3.42QNNRR1333 pKa = 11.84TVVKK1337 pKa = 10.59KK1338 pKa = 10.62LVLKK1342 pKa = 10.93GKK1344 pKa = 10.05
Molecular weight: 144.37 kDa
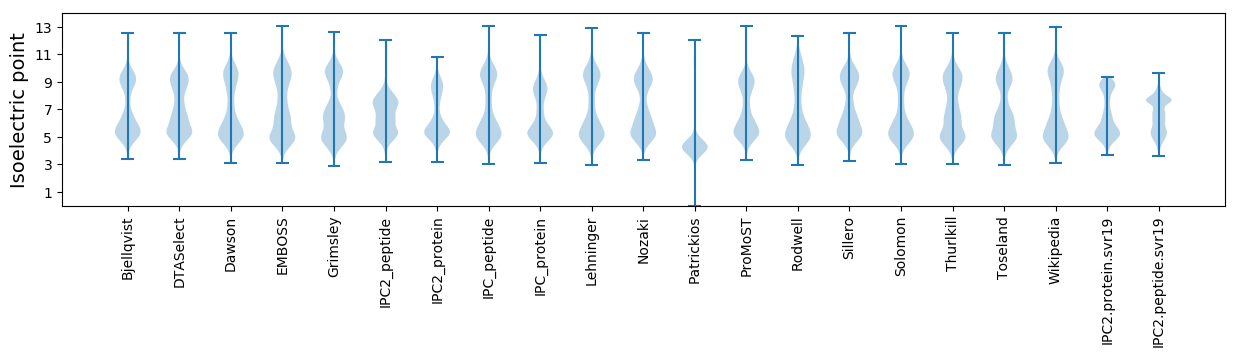
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5DP20|A0A1M5DP20_9BACT SPFH domain / Band 7 family protein OS=Mariniphaga anaerophila OX=1484053 GN=SAMN05444274_107184 PE=4 SV=1
MM1 pKa = 7.66ISNIQKK7 pKa = 10.23NLIRR11 pKa = 11.84TLTLQKK17 pKa = 10.01RR18 pKa = 11.84AKK20 pKa = 9.3KK21 pKa = 9.65RR22 pKa = 11.84RR23 pKa = 11.84LIGRR27 pKa = 11.84LFSQKK32 pKa = 10.51KK33 pKa = 9.38FLFNLKK39 pKa = 10.07SSPHH43 pKa = 5.05TLSLRR48 pKa = 11.84FQQHH52 pKa = 4.79RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84GQNPIFFGLIAQMDD69 pKa = 3.76
MM1 pKa = 7.66ISNIQKK7 pKa = 10.23NLIRR11 pKa = 11.84TLTLQKK17 pKa = 10.01RR18 pKa = 11.84AKK20 pKa = 9.3KK21 pKa = 9.65RR22 pKa = 11.84RR23 pKa = 11.84LIGRR27 pKa = 11.84LFSQKK32 pKa = 10.51KK33 pKa = 9.38FLFNLKK39 pKa = 10.07SSPHH43 pKa = 5.05TLSLRR48 pKa = 11.84FQQHH52 pKa = 4.79RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84GQNPIFFGLIAQMDD69 pKa = 3.76
Molecular weight: 8.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1523357 |
26 |
4578 |
383.3 |
43.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.623 ± 0.036 | 0.835 ± 0.016 |
5.36 ± 0.031 | 6.846 ± 0.034 |
5.292 ± 0.031 | 7.021 ± 0.036 |
1.76 ± 0.017 | 7.028 ± 0.03 |
6.954 ± 0.043 | 9.015 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.022 | 5.755 ± 0.037 |
3.836 ± 0.023 | 3.373 ± 0.02 |
4.073 ± 0.026 | 6.484 ± 0.032 |
5.407 ± 0.045 | 6.587 ± 0.032 |
1.383 ± 0.016 | 4.083 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
