
Human papillomavirus 21
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 1
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
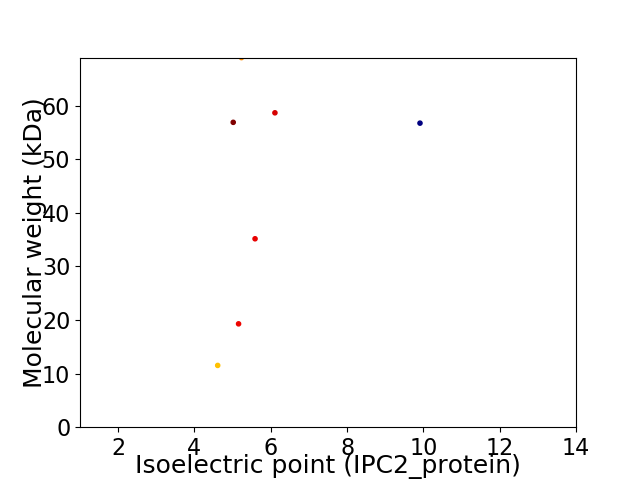
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P50787|VL1_HPV21 Major capsid protein L1 OS=Human papillomavirus 21 OX=31548 GN=L1 PE=3 SV=1
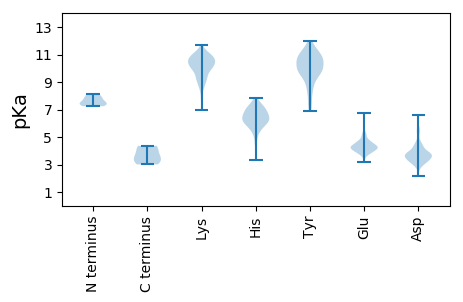
MM1 pKa = 7.37IGKK4 pKa = 8.93EE5 pKa = 3.9VTLQDD10 pKa = 3.25IVLEE14 pKa = 4.25LNEE17 pKa = 4.72LQPEE21 pKa = 4.3VQPVDD26 pKa = 3.84LFCEE30 pKa = 4.21EE31 pKa = 4.7EE32 pKa = 4.49LPSEE36 pKa = 4.09QQEE39 pKa = 4.58TEE41 pKa = 3.94EE42 pKa = 4.17EE43 pKa = 4.28LPEE46 pKa = 3.82RR47 pKa = 11.84TAYY50 pKa = 9.76KK51 pKa = 10.33VVTPCGCCKK60 pKa = 9.89VKK62 pKa = 10.76LRR64 pKa = 11.84IFVNATQFAIRR75 pKa = 11.84TFQNLLFEE83 pKa = 4.86EE84 pKa = 4.56LQLLCPEE91 pKa = 4.3CRR93 pKa = 11.84GNCKK97 pKa = 10.3HH98 pKa = 6.48GGSS101 pKa = 3.89
MM1 pKa = 7.37IGKK4 pKa = 8.93EE5 pKa = 3.9VTLQDD10 pKa = 3.25IVLEE14 pKa = 4.25LNEE17 pKa = 4.72LQPEE21 pKa = 4.3VQPVDD26 pKa = 3.84LFCEE30 pKa = 4.21EE31 pKa = 4.7EE32 pKa = 4.49LPSEE36 pKa = 4.09QQEE39 pKa = 4.58TEE41 pKa = 3.94EE42 pKa = 4.17EE43 pKa = 4.28LPEE46 pKa = 3.82RR47 pKa = 11.84TAYY50 pKa = 9.76KK51 pKa = 10.33VVTPCGCCKK60 pKa = 9.89VKK62 pKa = 10.76LRR64 pKa = 11.84IFVNATQFAIRR75 pKa = 11.84TFQNLLFEE83 pKa = 4.86EE84 pKa = 4.56LQLLCPEE91 pKa = 4.3CRR93 pKa = 11.84GNCKK97 pKa = 10.3HH98 pKa = 6.48GGSS101 pKa = 3.89
Molecular weight: 11.53 kDa
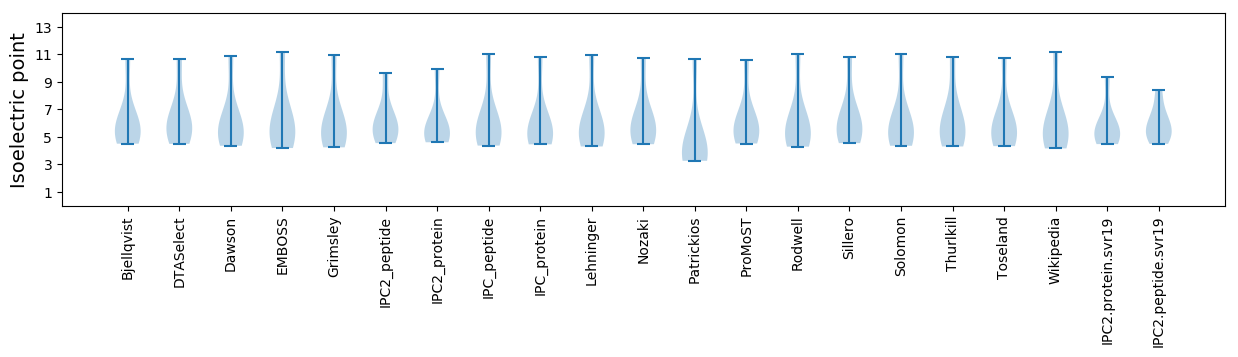
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P50779|VE7_HPV21 Protein E7 OS=Human papillomavirus 21 OX=31548 GN=E7 PE=3 SV=1
MM1 pKa = 7.73EE2 pKa = 5.02NLSDD6 pKa = 4.25RR7 pKa = 11.84FNVLQDD13 pKa = 3.28QLMNIYY19 pKa = 9.96EE20 pKa = 4.37SAANTIEE27 pKa = 3.99SQIEE31 pKa = 3.68HH32 pKa = 6.21WQTLRR37 pKa = 11.84KK38 pKa = 9.15EE39 pKa = 4.05AVLLYY44 pKa = 8.98FARR47 pKa = 11.84QKK49 pKa = 11.08GVTRR53 pKa = 11.84LGYY56 pKa = 10.42QYY58 pKa = 11.01VPPLAVSEE66 pKa = 4.48SRR68 pKa = 11.84AKK70 pKa = 10.0QAIGMMLQLQSLQKK84 pKa = 10.18SEE86 pKa = 4.19YY87 pKa = 10.66AKK89 pKa = 10.59EE90 pKa = 3.82PWSLVDD96 pKa = 3.61TSAEE100 pKa = 4.25TFRR103 pKa = 11.84SPPEE107 pKa = 3.58NHH109 pKa = 6.22FKK111 pKa = 10.7KK112 pKa = 10.69GPVSVEE118 pKa = 3.72VIYY121 pKa = 11.24DD122 pKa = 3.62NDD124 pKa = 3.68KK125 pKa = 11.71DD126 pKa = 3.79NANAYY131 pKa = 6.88TMWRR135 pKa = 11.84YY136 pKa = 10.06VYY138 pKa = 10.84YY139 pKa = 10.87VDD141 pKa = 6.59DD142 pKa = 5.43DD143 pKa = 4.53DD144 pKa = 4.44QWHH147 pKa = 6.9KK148 pKa = 11.3SPSGVNHH155 pKa = 5.96TGIYY159 pKa = 10.08FMQGTFRR166 pKa = 11.84HH167 pKa = 6.06YY168 pKa = 10.72YY169 pKa = 10.34VLFADD174 pKa = 4.09DD175 pKa = 3.6ASRR178 pKa = 11.84YY179 pKa = 9.65SRR181 pKa = 11.84TGHH184 pKa = 6.19WEE186 pKa = 3.84VNVNKK191 pKa = 8.65EE192 pKa = 4.3TVFAPVTSSTPPDD205 pKa = 3.8SPGGQADD212 pKa = 4.3SNTSSTTPATTTDD225 pKa = 3.11STSRR229 pKa = 11.84LSSTRR234 pKa = 11.84KK235 pKa = 9.57QSQQTNTKK243 pKa = 7.71GRR245 pKa = 11.84RR246 pKa = 11.84YY247 pKa = 9.84GRR249 pKa = 11.84RR250 pKa = 11.84PSSRR254 pKa = 11.84TRR256 pKa = 11.84RR257 pKa = 11.84TTQTHH262 pKa = 3.33QRR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84SRR268 pKa = 11.84SKK270 pKa = 9.28SRR272 pKa = 11.84SRR274 pKa = 11.84SRR276 pKa = 11.84SRR278 pKa = 11.84SRR280 pKa = 11.84LRR282 pKa = 11.84SRR284 pKa = 11.84SRR286 pKa = 11.84SRR288 pKa = 11.84SRR290 pKa = 11.84SYY292 pKa = 10.97SRR294 pKa = 11.84SRR296 pKa = 11.84SQSSDD301 pKa = 2.85QPQYY305 pKa = 10.72RR306 pKa = 11.84FRR308 pKa = 11.84SGGQVSLITTATTTTTTATNYY329 pKa = 6.89STRR332 pKa = 11.84GSGRR336 pKa = 11.84GSSSTSSSTSKK347 pKa = 10.38RR348 pKa = 11.84PRR350 pKa = 11.84RR351 pKa = 11.84PRR353 pKa = 11.84GGAIGGSSGRR363 pKa = 11.84GRR365 pKa = 11.84RR366 pKa = 11.84SSSTSPSPSKK376 pKa = 10.44RR377 pKa = 11.84SRR379 pKa = 11.84GKK381 pKa = 10.25SEE383 pKa = 3.94SVRR386 pKa = 11.84QRR388 pKa = 11.84GISPDD393 pKa = 3.65DD394 pKa = 3.59VGKK397 pKa = 10.53SLQSVSTRR405 pKa = 11.84NTGRR409 pKa = 11.84LGRR412 pKa = 11.84LLDD415 pKa = 3.94EE416 pKa = 5.3ALDD419 pKa = 3.88PPVILVRR426 pKa = 11.84GEE428 pKa = 4.16PNTLKK433 pKa = 10.67CFRR436 pKa = 11.84NRR438 pKa = 11.84AKK440 pKa = 10.61LKK442 pKa = 10.33YY443 pKa = 9.64AGLYY447 pKa = 9.9KK448 pKa = 10.58AFSTAWSWVAGDD460 pKa = 3.33GTEE463 pKa = 3.75RR464 pKa = 11.84LGRR467 pKa = 11.84SRR469 pKa = 11.84MLISFFSFEE478 pKa = 3.86QRR480 pKa = 11.84KK481 pKa = 10.04DD482 pKa = 2.92FDD484 pKa = 3.77KK485 pKa = 10.79TVKK488 pKa = 10.16YY489 pKa = 9.85PKK491 pKa = 10.59GVDD494 pKa = 2.96RR495 pKa = 11.84SYY497 pKa = 12.02GSFDD501 pKa = 3.4SLL503 pKa = 4.36
MM1 pKa = 7.73EE2 pKa = 5.02NLSDD6 pKa = 4.25RR7 pKa = 11.84FNVLQDD13 pKa = 3.28QLMNIYY19 pKa = 9.96EE20 pKa = 4.37SAANTIEE27 pKa = 3.99SQIEE31 pKa = 3.68HH32 pKa = 6.21WQTLRR37 pKa = 11.84KK38 pKa = 9.15EE39 pKa = 4.05AVLLYY44 pKa = 8.98FARR47 pKa = 11.84QKK49 pKa = 11.08GVTRR53 pKa = 11.84LGYY56 pKa = 10.42QYY58 pKa = 11.01VPPLAVSEE66 pKa = 4.48SRR68 pKa = 11.84AKK70 pKa = 10.0QAIGMMLQLQSLQKK84 pKa = 10.18SEE86 pKa = 4.19YY87 pKa = 10.66AKK89 pKa = 10.59EE90 pKa = 3.82PWSLVDD96 pKa = 3.61TSAEE100 pKa = 4.25TFRR103 pKa = 11.84SPPEE107 pKa = 3.58NHH109 pKa = 6.22FKK111 pKa = 10.7KK112 pKa = 10.69GPVSVEE118 pKa = 3.72VIYY121 pKa = 11.24DD122 pKa = 3.62NDD124 pKa = 3.68KK125 pKa = 11.71DD126 pKa = 3.79NANAYY131 pKa = 6.88TMWRR135 pKa = 11.84YY136 pKa = 10.06VYY138 pKa = 10.84YY139 pKa = 10.87VDD141 pKa = 6.59DD142 pKa = 5.43DD143 pKa = 4.53DD144 pKa = 4.44QWHH147 pKa = 6.9KK148 pKa = 11.3SPSGVNHH155 pKa = 5.96TGIYY159 pKa = 10.08FMQGTFRR166 pKa = 11.84HH167 pKa = 6.06YY168 pKa = 10.72YY169 pKa = 10.34VLFADD174 pKa = 4.09DD175 pKa = 3.6ASRR178 pKa = 11.84YY179 pKa = 9.65SRR181 pKa = 11.84TGHH184 pKa = 6.19WEE186 pKa = 3.84VNVNKK191 pKa = 8.65EE192 pKa = 4.3TVFAPVTSSTPPDD205 pKa = 3.8SPGGQADD212 pKa = 4.3SNTSSTTPATTTDD225 pKa = 3.11STSRR229 pKa = 11.84LSSTRR234 pKa = 11.84KK235 pKa = 9.57QSQQTNTKK243 pKa = 7.71GRR245 pKa = 11.84RR246 pKa = 11.84YY247 pKa = 9.84GRR249 pKa = 11.84RR250 pKa = 11.84PSSRR254 pKa = 11.84TRR256 pKa = 11.84RR257 pKa = 11.84TTQTHH262 pKa = 3.33QRR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84SRR268 pKa = 11.84SKK270 pKa = 9.28SRR272 pKa = 11.84SRR274 pKa = 11.84SRR276 pKa = 11.84SRR278 pKa = 11.84SRR280 pKa = 11.84LRR282 pKa = 11.84SRR284 pKa = 11.84SRR286 pKa = 11.84SRR288 pKa = 11.84SRR290 pKa = 11.84SYY292 pKa = 10.97SRR294 pKa = 11.84SRR296 pKa = 11.84SQSSDD301 pKa = 2.85QPQYY305 pKa = 10.72RR306 pKa = 11.84FRR308 pKa = 11.84SGGQVSLITTATTTTTTATNYY329 pKa = 6.89STRR332 pKa = 11.84GSGRR336 pKa = 11.84GSSSTSSSTSKK347 pKa = 10.38RR348 pKa = 11.84PRR350 pKa = 11.84RR351 pKa = 11.84PRR353 pKa = 11.84GGAIGGSSGRR363 pKa = 11.84GRR365 pKa = 11.84RR366 pKa = 11.84SSSTSPSPSKK376 pKa = 10.44RR377 pKa = 11.84SRR379 pKa = 11.84GKK381 pKa = 10.25SEE383 pKa = 3.94SVRR386 pKa = 11.84QRR388 pKa = 11.84GISPDD393 pKa = 3.65DD394 pKa = 3.59VGKK397 pKa = 10.53SLQSVSTRR405 pKa = 11.84NTGRR409 pKa = 11.84LGRR412 pKa = 11.84LLDD415 pKa = 3.94EE416 pKa = 5.3ALDD419 pKa = 3.88PPVILVRR426 pKa = 11.84GEE428 pKa = 4.16PNTLKK433 pKa = 10.67CFRR436 pKa = 11.84NRR438 pKa = 11.84AKK440 pKa = 10.61LKK442 pKa = 10.33YY443 pKa = 9.64AGLYY447 pKa = 9.9KK448 pKa = 10.58AFSTAWSWVAGDD460 pKa = 3.33GTEE463 pKa = 3.75RR464 pKa = 11.84LGRR467 pKa = 11.84SRR469 pKa = 11.84MLISFFSFEE478 pKa = 3.86QRR480 pKa = 11.84KK481 pKa = 10.04DD482 pKa = 2.92FDD484 pKa = 3.77KK485 pKa = 10.79TVKK488 pKa = 10.16YY489 pKa = 9.85PKK491 pKa = 10.59GVDD494 pKa = 2.96RR495 pKa = 11.84SYY497 pKa = 12.02GSFDD501 pKa = 3.4SLL503 pKa = 4.36
Molecular weight: 56.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2739 |
101 |
603 |
391.3 |
43.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.111 ± 0.656 | 1.899 ± 0.624 |
5.988 ± 0.345 | 6.17 ± 0.891 |
3.943 ± 0.481 | 6.645 ± 0.769 |
2.081 ± 0.261 | 4.527 ± 0.529 |
4.892 ± 0.549 | 7.959 ± 0.761 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.789 ± 0.398 | 4.381 ± 0.666 |
7.375 ± 1.798 | 4.856 ± 0.421 |
6.608 ± 1.165 | 8.58 ± 1.421 |
6.937 ± 1.12 | 5.769 ± 0.517 |
1.168 ± 0.288 | 3.322 ± 0.48 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
