
Muromegalovirus G4
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Muromegalovirus; Murid betaherpesvirus 1
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
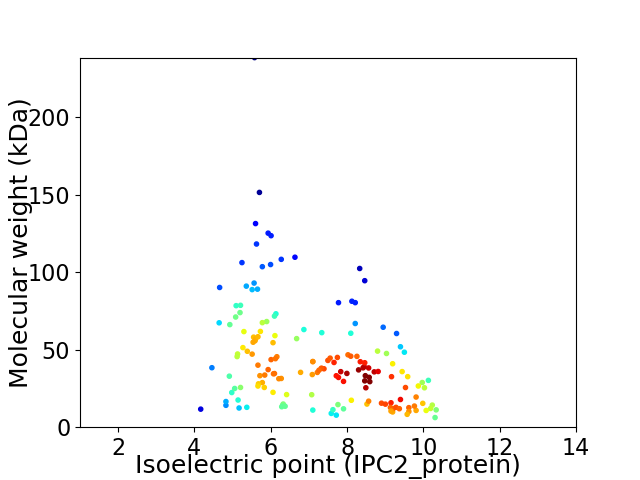
Virtual 2D-PAGE plot for 163 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B3UX83|B3UX83_MUHV1 M138 OS=Muromegalovirus G4 OX=524650 GN=m138 PE=4 SV=1
MM1 pKa = 7.92AINTTEE7 pKa = 4.23CPSPSEE13 pKa = 4.15TVSAQTIVLTCAALTSVLLAAAILVVAISALVRR46 pKa = 11.84RR47 pKa = 11.84CRR49 pKa = 11.84RR50 pKa = 11.84QQTPRR55 pKa = 11.84GDD57 pKa = 3.38EE58 pKa = 3.82QLPIEE63 pKa = 4.42CSVYY67 pKa = 10.08LVQNNGVDD75 pKa = 4.75DD76 pKa = 5.38DD77 pKa = 5.51DD78 pKa = 4.77IPPGFLGRR86 pKa = 11.84PAGYY90 pKa = 10.42EE91 pKa = 3.8DD92 pKa = 4.46MSPHH96 pKa = 6.63SFPLDD101 pKa = 3.9EE102 pKa = 5.83IYY104 pKa = 11.02EE105 pKa = 4.53SIDD108 pKa = 3.11
MM1 pKa = 7.92AINTTEE7 pKa = 4.23CPSPSEE13 pKa = 4.15TVSAQTIVLTCAALTSVLLAAAILVVAISALVRR46 pKa = 11.84RR47 pKa = 11.84CRR49 pKa = 11.84RR50 pKa = 11.84QQTPRR55 pKa = 11.84GDD57 pKa = 3.38EE58 pKa = 3.82QLPIEE63 pKa = 4.42CSVYY67 pKa = 10.08LVQNNGVDD75 pKa = 4.75DD76 pKa = 5.38DD77 pKa = 5.51DD78 pKa = 4.77IPPGFLGRR86 pKa = 11.84PAGYY90 pKa = 10.42EE91 pKa = 3.8DD92 pKa = 4.46MSPHH96 pKa = 6.63SFPLDD101 pKa = 3.9EE102 pKa = 5.83IYY104 pKa = 11.02EE105 pKa = 4.53SIDD108 pKa = 3.11
Molecular weight: 11.65 kDa
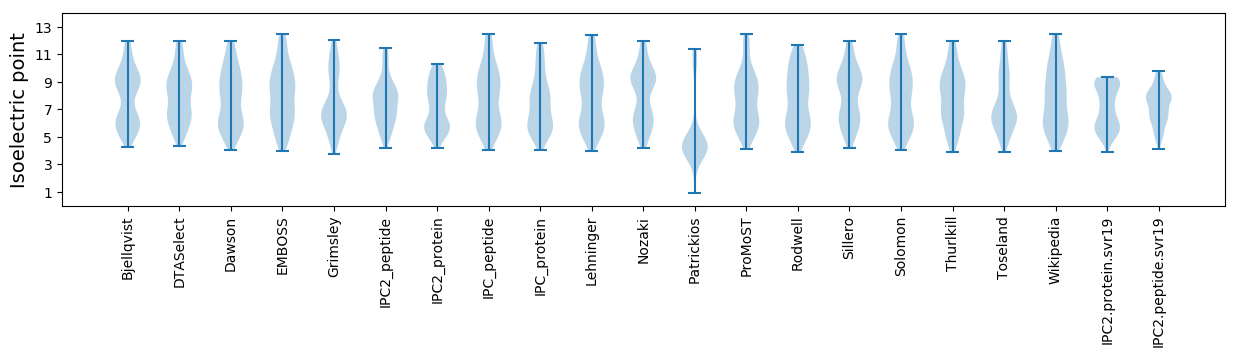
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B3UWX9|B3UWX9_MUHV1 M26 OS=Muromegalovirus G4 OX=524650 GN=M26 PE=4 SV=1
MM1 pKa = 7.61PASSCTSRR9 pKa = 11.84TTSRR13 pKa = 11.84SSSRR17 pKa = 11.84TASLEE22 pKa = 3.79GTSRR26 pKa = 11.84YY27 pKa = 9.99SATCVRR33 pKa = 11.84ASYY36 pKa = 10.27RR37 pKa = 11.84RR38 pKa = 11.84SSRR41 pKa = 11.84SASATSRR48 pKa = 11.84VLPGYY53 pKa = 9.6RR54 pKa = 11.84RR55 pKa = 11.84PSASGCPSSSEE66 pKa = 4.53AYY68 pKa = 9.32SAPGSRR74 pKa = 11.84PRR76 pKa = 11.84GSAGRR81 pKa = 11.84SDD83 pKa = 3.64SVPRR87 pKa = 11.84IFRR90 pKa = 11.84TGCLRR95 pKa = 11.84RR96 pKa = 11.84PWLCPTCEE104 pKa = 3.81RR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84PPLRR113 pKa = 11.84PWRR116 pKa = 11.84PQRR119 pKa = 11.84RR120 pKa = 11.84SDD122 pKa = 3.08TVARR126 pKa = 11.84KK127 pKa = 7.55RR128 pKa = 11.84TRR130 pKa = 11.84SRR132 pKa = 11.84TRR134 pKa = 11.84TRR136 pKa = 11.84TRR138 pKa = 11.84IRR140 pKa = 11.84NGAKK144 pKa = 10.04GDD146 pKa = 3.69QTMLAPAAAVSMPEE160 pKa = 4.16SEE162 pKa = 4.4TEE164 pKa = 4.03MVTVVPARR172 pKa = 11.84STLAAKK178 pKa = 9.64AAAMRR183 pKa = 11.84KK184 pKa = 9.08AYY186 pKa = 10.05AVNRR190 pKa = 11.84LLALGRR196 pKa = 11.84AAAEE200 pKa = 4.4GKK202 pKa = 10.26LEE204 pKa = 3.88QWLSARR210 pKa = 11.84RR211 pKa = 11.84GEE213 pKa = 4.48RR214 pKa = 11.84IPLVFPEE221 pKa = 4.44KK222 pKa = 9.78CAKK225 pKa = 10.26YY226 pKa = 10.48LILCAPGDD234 pKa = 4.08VDD236 pKa = 5.18GHH238 pKa = 6.02EE239 pKa = 5.07CDD241 pKa = 4.15PDD243 pKa = 3.47ALRR246 pKa = 11.84ALEE249 pKa = 4.12PAVDD253 pKa = 3.61RR254 pKa = 11.84LVVVGFVEE262 pKa = 4.37GRR264 pKa = 11.84EE265 pKa = 3.97DD266 pKa = 3.18AHH268 pKa = 8.0RR269 pKa = 11.84DD270 pKa = 3.14GDD272 pKa = 3.9DD273 pKa = 3.28RR274 pKa = 11.84CIVLVNPSSAVFIYY288 pKa = 10.67DD289 pKa = 3.59PAPYY293 pKa = 10.2GGLYY297 pKa = 10.35RR298 pKa = 11.84LANTMIGFVRR308 pKa = 11.84RR309 pKa = 11.84GLRR312 pKa = 11.84RR313 pKa = 11.84FDD315 pKa = 3.68SIYY318 pKa = 10.42RR319 pKa = 11.84DD320 pKa = 3.69PCCVEE325 pKa = 3.52DD326 pKa = 5.17AIRR329 pKa = 11.84VKK331 pKa = 10.62DD332 pKa = 3.59GHH334 pKa = 6.96ASLPRR339 pKa = 11.84NADD342 pKa = 3.23EE343 pKa = 4.95ALAAAEE349 pKa = 4.49RR350 pKa = 11.84GLACTLQWPEE360 pKa = 4.61DD361 pKa = 3.77YY362 pKa = 11.05DD363 pKa = 4.35FVFGTPPEE371 pKa = 4.23SSNAVASSLNKK382 pKa = 9.6EE383 pKa = 3.83QSPCIDD389 pKa = 3.64ALRR392 pKa = 11.84LSTFGHH398 pKa = 6.75FGTVAGLSSTRR409 pKa = 11.84PRR411 pKa = 11.84TAVFIGADD419 pKa = 3.27HH420 pKa = 6.65AVYY423 pKa = 10.15AIHH426 pKa = 6.75HH427 pKa = 6.31AVSKK431 pKa = 10.27LLRR434 pKa = 11.84LAEE437 pKa = 4.32SLDD440 pKa = 3.45MFYY443 pKa = 10.9KK444 pKa = 10.3IGIRR448 pKa = 11.84RR449 pKa = 11.84FFHH452 pKa = 6.75NYY454 pKa = 8.12RR455 pKa = 11.84VVPSRR460 pKa = 11.84CGDD463 pKa = 3.13DD464 pKa = 4.07RR465 pKa = 11.84FFLKK469 pKa = 10.65AVV471 pKa = 3.35
MM1 pKa = 7.61PASSCTSRR9 pKa = 11.84TTSRR13 pKa = 11.84SSSRR17 pKa = 11.84TASLEE22 pKa = 3.79GTSRR26 pKa = 11.84YY27 pKa = 9.99SATCVRR33 pKa = 11.84ASYY36 pKa = 10.27RR37 pKa = 11.84RR38 pKa = 11.84SSRR41 pKa = 11.84SASATSRR48 pKa = 11.84VLPGYY53 pKa = 9.6RR54 pKa = 11.84RR55 pKa = 11.84PSASGCPSSSEE66 pKa = 4.53AYY68 pKa = 9.32SAPGSRR74 pKa = 11.84PRR76 pKa = 11.84GSAGRR81 pKa = 11.84SDD83 pKa = 3.64SVPRR87 pKa = 11.84IFRR90 pKa = 11.84TGCLRR95 pKa = 11.84RR96 pKa = 11.84PWLCPTCEE104 pKa = 3.81RR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84PPLRR113 pKa = 11.84PWRR116 pKa = 11.84PQRR119 pKa = 11.84RR120 pKa = 11.84SDD122 pKa = 3.08TVARR126 pKa = 11.84KK127 pKa = 7.55RR128 pKa = 11.84TRR130 pKa = 11.84SRR132 pKa = 11.84TRR134 pKa = 11.84TRR136 pKa = 11.84TRR138 pKa = 11.84IRR140 pKa = 11.84NGAKK144 pKa = 10.04GDD146 pKa = 3.69QTMLAPAAAVSMPEE160 pKa = 4.16SEE162 pKa = 4.4TEE164 pKa = 4.03MVTVVPARR172 pKa = 11.84STLAAKK178 pKa = 9.64AAAMRR183 pKa = 11.84KK184 pKa = 9.08AYY186 pKa = 10.05AVNRR190 pKa = 11.84LLALGRR196 pKa = 11.84AAAEE200 pKa = 4.4GKK202 pKa = 10.26LEE204 pKa = 3.88QWLSARR210 pKa = 11.84RR211 pKa = 11.84GEE213 pKa = 4.48RR214 pKa = 11.84IPLVFPEE221 pKa = 4.44KK222 pKa = 9.78CAKK225 pKa = 10.26YY226 pKa = 10.48LILCAPGDD234 pKa = 4.08VDD236 pKa = 5.18GHH238 pKa = 6.02EE239 pKa = 5.07CDD241 pKa = 4.15PDD243 pKa = 3.47ALRR246 pKa = 11.84ALEE249 pKa = 4.12PAVDD253 pKa = 3.61RR254 pKa = 11.84LVVVGFVEE262 pKa = 4.37GRR264 pKa = 11.84EE265 pKa = 3.97DD266 pKa = 3.18AHH268 pKa = 8.0RR269 pKa = 11.84DD270 pKa = 3.14GDD272 pKa = 3.9DD273 pKa = 3.28RR274 pKa = 11.84CIVLVNPSSAVFIYY288 pKa = 10.67DD289 pKa = 3.59PAPYY293 pKa = 10.2GGLYY297 pKa = 10.35RR298 pKa = 11.84LANTMIGFVRR308 pKa = 11.84RR309 pKa = 11.84GLRR312 pKa = 11.84RR313 pKa = 11.84FDD315 pKa = 3.68SIYY318 pKa = 10.42RR319 pKa = 11.84DD320 pKa = 3.69PCCVEE325 pKa = 3.52DD326 pKa = 5.17AIRR329 pKa = 11.84VKK331 pKa = 10.62DD332 pKa = 3.59GHH334 pKa = 6.96ASLPRR339 pKa = 11.84NADD342 pKa = 3.23EE343 pKa = 4.95ALAAAEE349 pKa = 4.49RR350 pKa = 11.84GLACTLQWPEE360 pKa = 4.61DD361 pKa = 3.77YY362 pKa = 11.05DD363 pKa = 4.35FVFGTPPEE371 pKa = 4.23SSNAVASSLNKK382 pKa = 9.6EE383 pKa = 3.83QSPCIDD389 pKa = 3.64ALRR392 pKa = 11.84LSTFGHH398 pKa = 6.75FGTVAGLSSTRR409 pKa = 11.84PRR411 pKa = 11.84TAVFIGADD419 pKa = 3.27HH420 pKa = 6.65AVYY423 pKa = 10.15AIHH426 pKa = 6.75HH427 pKa = 6.31AVSKK431 pKa = 10.27LLRR434 pKa = 11.84LAEE437 pKa = 4.32SLDD440 pKa = 3.45MFYY443 pKa = 10.9KK444 pKa = 10.3IGIRR448 pKa = 11.84RR449 pKa = 11.84FFHH452 pKa = 6.75NYY454 pKa = 8.12RR455 pKa = 11.84VVPSRR460 pKa = 11.84CGDD463 pKa = 3.13DD464 pKa = 4.07RR465 pKa = 11.84FFLKK469 pKa = 10.65AVV471 pKa = 3.35
Molecular weight: 51.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
65346 |
57 |
2149 |
400.9 |
44.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.036 ± 0.171 | 2.207 ± 0.094 |
5.893 ± 0.146 | 5.356 ± 0.142 |
3.791 ± 0.114 | 6.635 ± 0.225 |
2.208 ± 0.098 | 4.358 ± 0.115 |
3.506 ± 0.133 | 8.524 ± 0.181 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.074 | 3.298 ± 0.09 |
6.034 ± 0.204 | 2.911 ± 0.105 |
8.118 ± 0.223 | 8.565 ± 0.242 |
6.894 ± 0.241 | 7.428 ± 0.159 |
1.065 ± 0.064 | 2.975 ± 0.099 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
