
Shahe narna-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
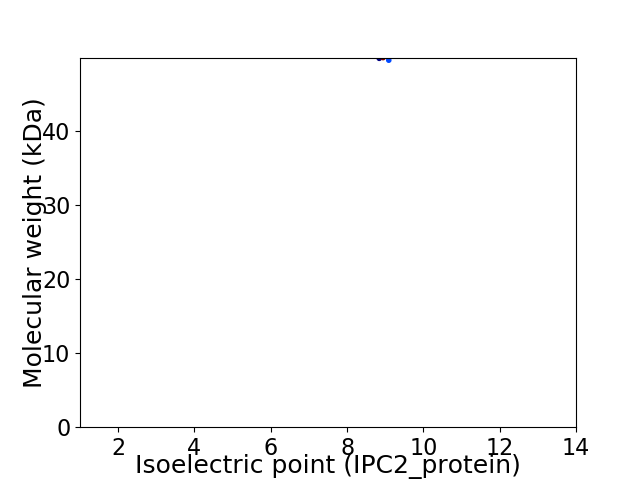
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KI92|A0A1L3KI92_9VIRU RNA-dependent RNA polymerase OS=Shahe narna-like virus 5 OX=1923433 PE=4 SV=1
MM1 pKa = 7.8KK2 pKa = 9.57MFKK5 pKa = 10.59YY6 pKa = 10.01MRR8 pKa = 11.84GDD10 pKa = 3.56ATWDD14 pKa = 3.1QEE16 pKa = 4.13KK17 pKa = 9.88QVRR20 pKa = 11.84RR21 pKa = 11.84ICKK24 pKa = 6.41WTEE27 pKa = 3.32RR28 pKa = 11.84GMKK31 pKa = 9.99LYY33 pKa = 11.02SFDD36 pKa = 3.35LTAATDD42 pKa = 3.74RR43 pKa = 11.84WPAWHH48 pKa = 6.37QKK50 pKa = 10.33LVVDD54 pKa = 4.28RR55 pKa = 11.84VFNKK59 pKa = 10.8GIGEE63 pKa = 3.87MWFRR67 pKa = 11.84LMTTTTPYY75 pKa = 9.35ATEE78 pKa = 4.05LKK80 pKa = 10.28RR81 pKa = 11.84YY82 pKa = 8.36IRR84 pKa = 11.84YY85 pKa = 9.86AIGQPMGAYY94 pKa = 10.19SSWAVLNMTHH104 pKa = 6.87HH105 pKa = 5.5YY106 pKa = 10.15VIRR109 pKa = 11.84LCAKK113 pKa = 10.16RR114 pKa = 11.84IGVKK118 pKa = 10.16ALYY121 pKa = 10.13SVLGDD126 pKa = 3.51DD127 pKa = 3.85VVIVGKK133 pKa = 10.11EE134 pKa = 3.5LADD137 pKa = 3.85EE138 pKa = 4.1YY139 pKa = 10.96RR140 pKa = 11.84SYY142 pKa = 9.78ITRR145 pKa = 11.84LGVKK149 pKa = 10.26LNLNKK154 pKa = 10.56SILNTDD160 pKa = 4.88FGPCSAEE167 pKa = 3.9FARR170 pKa = 11.84HH171 pKa = 4.76IVRR174 pKa = 11.84DD175 pKa = 3.47GHH177 pKa = 6.84IVGTFSPSLVRR188 pKa = 11.84NIYY191 pKa = 10.53SYY193 pKa = 10.33TDD195 pKa = 3.19VYY197 pKa = 11.69ASIDD201 pKa = 3.63LLRR204 pKa = 11.84EE205 pKa = 4.12VQSKK209 pKa = 10.08LGQVIHH215 pKa = 6.84VYY217 pKa = 10.81EE218 pKa = 4.39NTTLLPIPIVRR229 pKa = 11.84LLKK232 pKa = 9.98TFSFKK237 pKa = 10.93DD238 pKa = 3.33DD239 pKa = 3.61LLAIVTVPLTALKK252 pKa = 10.38LPRR255 pKa = 11.84VYY257 pKa = 10.47HH258 pKa = 5.59PEE260 pKa = 3.92KK261 pKa = 10.9FEE263 pKa = 4.25GDD265 pKa = 3.51PMVGYY270 pKa = 10.73DD271 pKa = 3.52KK272 pKa = 10.83FANPWYY278 pKa = 9.47EE279 pKa = 3.73CDD281 pKa = 3.42INSVNAEE288 pKa = 4.28FGNKK292 pKa = 7.03WMRR295 pKa = 11.84EE296 pKa = 3.4ASLRR300 pKa = 11.84MNKK303 pKa = 10.02LEE305 pKa = 4.32SLLDD309 pKa = 3.57KK310 pKa = 11.24LGDD313 pKa = 4.01GGSTVYY319 pKa = 10.32RR320 pKa = 11.84GRR322 pKa = 11.84LLEE325 pKa = 4.32ISSHH329 pKa = 5.11PLRR332 pKa = 11.84YY333 pKa = 9.47SLKK336 pKa = 8.63PLKK339 pKa = 10.23EE340 pKa = 3.8EE341 pKa = 5.21LMNQFWRR348 pKa = 11.84VMRR351 pKa = 11.84GEE353 pKa = 4.1EE354 pKa = 4.05VPIFSSQLQTDD365 pKa = 3.22IDD367 pKa = 4.22LLIFTLTEE375 pKa = 4.08GKK377 pKa = 9.91SFRR380 pKa = 11.84TWRR383 pKa = 11.84NSKK386 pKa = 7.86TSKK389 pKa = 10.14RR390 pKa = 11.84KK391 pKa = 8.1HH392 pKa = 5.21ACMLIKK398 pKa = 10.35KK399 pKa = 8.04VLKK402 pKa = 9.81EE403 pKa = 3.91SHH405 pKa = 6.03RR406 pKa = 11.84VAAAMKK412 pKa = 8.36EE413 pKa = 4.09LPTTSMSWEE422 pKa = 4.37DD423 pKa = 3.46YY424 pKa = 10.98AKK426 pKa = 10.51QFEE429 pKa = 4.31
MM1 pKa = 7.8KK2 pKa = 9.57MFKK5 pKa = 10.59YY6 pKa = 10.01MRR8 pKa = 11.84GDD10 pKa = 3.56ATWDD14 pKa = 3.1QEE16 pKa = 4.13KK17 pKa = 9.88QVRR20 pKa = 11.84RR21 pKa = 11.84ICKK24 pKa = 6.41WTEE27 pKa = 3.32RR28 pKa = 11.84GMKK31 pKa = 9.99LYY33 pKa = 11.02SFDD36 pKa = 3.35LTAATDD42 pKa = 3.74RR43 pKa = 11.84WPAWHH48 pKa = 6.37QKK50 pKa = 10.33LVVDD54 pKa = 4.28RR55 pKa = 11.84VFNKK59 pKa = 10.8GIGEE63 pKa = 3.87MWFRR67 pKa = 11.84LMTTTTPYY75 pKa = 9.35ATEE78 pKa = 4.05LKK80 pKa = 10.28RR81 pKa = 11.84YY82 pKa = 8.36IRR84 pKa = 11.84YY85 pKa = 9.86AIGQPMGAYY94 pKa = 10.19SSWAVLNMTHH104 pKa = 6.87HH105 pKa = 5.5YY106 pKa = 10.15VIRR109 pKa = 11.84LCAKK113 pKa = 10.16RR114 pKa = 11.84IGVKK118 pKa = 10.16ALYY121 pKa = 10.13SVLGDD126 pKa = 3.51DD127 pKa = 3.85VVIVGKK133 pKa = 10.11EE134 pKa = 3.5LADD137 pKa = 3.85EE138 pKa = 4.1YY139 pKa = 10.96RR140 pKa = 11.84SYY142 pKa = 9.78ITRR145 pKa = 11.84LGVKK149 pKa = 10.26LNLNKK154 pKa = 10.56SILNTDD160 pKa = 4.88FGPCSAEE167 pKa = 3.9FARR170 pKa = 11.84HH171 pKa = 4.76IVRR174 pKa = 11.84DD175 pKa = 3.47GHH177 pKa = 6.84IVGTFSPSLVRR188 pKa = 11.84NIYY191 pKa = 10.53SYY193 pKa = 10.33TDD195 pKa = 3.19VYY197 pKa = 11.69ASIDD201 pKa = 3.63LLRR204 pKa = 11.84EE205 pKa = 4.12VQSKK209 pKa = 10.08LGQVIHH215 pKa = 6.84VYY217 pKa = 10.81EE218 pKa = 4.39NTTLLPIPIVRR229 pKa = 11.84LLKK232 pKa = 9.98TFSFKK237 pKa = 10.93DD238 pKa = 3.33DD239 pKa = 3.61LLAIVTVPLTALKK252 pKa = 10.38LPRR255 pKa = 11.84VYY257 pKa = 10.47HH258 pKa = 5.59PEE260 pKa = 3.92KK261 pKa = 10.9FEE263 pKa = 4.25GDD265 pKa = 3.51PMVGYY270 pKa = 10.73DD271 pKa = 3.52KK272 pKa = 10.83FANPWYY278 pKa = 9.47EE279 pKa = 3.73CDD281 pKa = 3.42INSVNAEE288 pKa = 4.28FGNKK292 pKa = 7.03WMRR295 pKa = 11.84EE296 pKa = 3.4ASLRR300 pKa = 11.84MNKK303 pKa = 10.02LEE305 pKa = 4.32SLLDD309 pKa = 3.57KK310 pKa = 11.24LGDD313 pKa = 4.01GGSTVYY319 pKa = 10.32RR320 pKa = 11.84GRR322 pKa = 11.84LLEE325 pKa = 4.32ISSHH329 pKa = 5.11PLRR332 pKa = 11.84YY333 pKa = 9.47SLKK336 pKa = 8.63PLKK339 pKa = 10.23EE340 pKa = 3.8EE341 pKa = 5.21LMNQFWRR348 pKa = 11.84VMRR351 pKa = 11.84GEE353 pKa = 4.1EE354 pKa = 4.05VPIFSSQLQTDD365 pKa = 3.22IDD367 pKa = 4.22LLIFTLTEE375 pKa = 4.08GKK377 pKa = 9.91SFRR380 pKa = 11.84TWRR383 pKa = 11.84NSKK386 pKa = 7.86TSKK389 pKa = 10.14RR390 pKa = 11.84KK391 pKa = 8.1HH392 pKa = 5.21ACMLIKK398 pKa = 10.35KK399 pKa = 8.04VLKK402 pKa = 9.81EE403 pKa = 3.91SHH405 pKa = 6.03RR406 pKa = 11.84VAAAMKK412 pKa = 8.36EE413 pKa = 4.09LPTTSMSWEE422 pKa = 4.37DD423 pKa = 3.46YY424 pKa = 10.98AKK426 pKa = 10.51QFEE429 pKa = 4.31
Molecular weight: 49.78 kDa
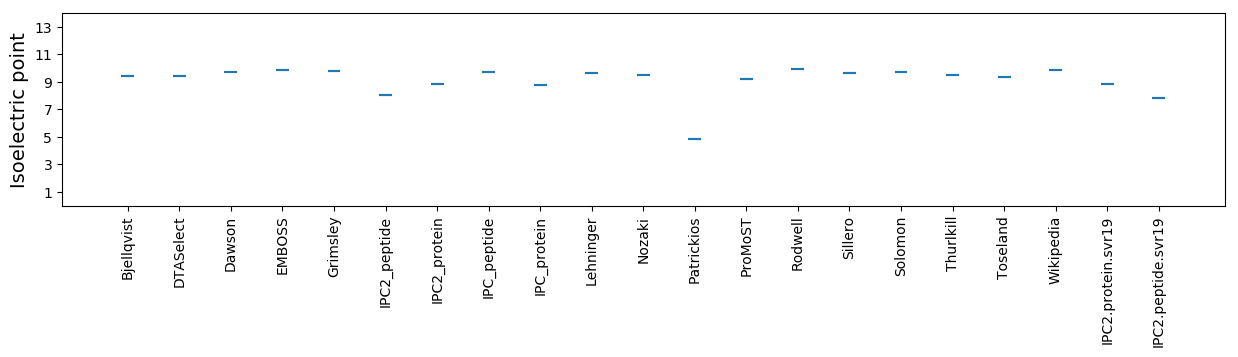
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI92|A0A1L3KI92_9VIRU RNA-dependent RNA polymerase OS=Shahe narna-like virus 5 OX=1923433 PE=4 SV=1
MM1 pKa = 7.8KK2 pKa = 9.57MFKK5 pKa = 10.59YY6 pKa = 10.01MRR8 pKa = 11.84GDD10 pKa = 3.56ATWDD14 pKa = 3.1QEE16 pKa = 4.13KK17 pKa = 9.88QVRR20 pKa = 11.84RR21 pKa = 11.84ICKK24 pKa = 6.41WTEE27 pKa = 3.32RR28 pKa = 11.84GMKK31 pKa = 9.99LYY33 pKa = 11.02SFDD36 pKa = 3.35LTAATDD42 pKa = 3.74RR43 pKa = 11.84WPAWHH48 pKa = 6.37QKK50 pKa = 10.33LVVDD54 pKa = 4.28RR55 pKa = 11.84VFNKK59 pKa = 10.8GIGEE63 pKa = 3.87MWFRR67 pKa = 11.84LMTTTTPYY75 pKa = 9.35ATEE78 pKa = 4.05LKK80 pKa = 10.28RR81 pKa = 11.84YY82 pKa = 8.36IRR84 pKa = 11.84YY85 pKa = 9.86AIGQPMGAYY94 pKa = 10.19SSWAVLNMTHH104 pKa = 6.87HH105 pKa = 5.5YY106 pKa = 10.15VIRR109 pKa = 11.84LCAKK113 pKa = 10.16RR114 pKa = 11.84IGVKK118 pKa = 10.16ALYY121 pKa = 10.13SVLGDD126 pKa = 3.51DD127 pKa = 3.85VVIVGKK133 pKa = 10.11EE134 pKa = 3.5LADD137 pKa = 3.85EE138 pKa = 4.1YY139 pKa = 10.96RR140 pKa = 11.84SYY142 pKa = 9.78ITRR145 pKa = 11.84LGVKK149 pKa = 10.26LNLNKK154 pKa = 10.56SILNTDD160 pKa = 4.88FGPCSAEE167 pKa = 3.9FARR170 pKa = 11.84HH171 pKa = 4.76IVRR174 pKa = 11.84DD175 pKa = 3.47GHH177 pKa = 6.84IVGTFSPSLVRR188 pKa = 11.84NIYY191 pKa = 10.53SYY193 pKa = 10.33TDD195 pKa = 3.19VYY197 pKa = 11.69ASIDD201 pKa = 3.63LLRR204 pKa = 11.84EE205 pKa = 4.12VQSKK209 pKa = 10.08LGQVIHH215 pKa = 6.84VYY217 pKa = 10.81EE218 pKa = 4.39NTTLLPIPIVRR229 pKa = 11.84LLKK232 pKa = 9.98TFSFKK237 pKa = 10.93DD238 pKa = 3.33DD239 pKa = 3.61LLAIVTVPLTALKK252 pKa = 10.38LPRR255 pKa = 11.84VYY257 pKa = 10.47HH258 pKa = 5.59PEE260 pKa = 3.92KK261 pKa = 10.9FEE263 pKa = 4.25GDD265 pKa = 3.51PMVGYY270 pKa = 10.73DD271 pKa = 3.52KK272 pKa = 10.83FANPWYY278 pKa = 9.47EE279 pKa = 3.73CDD281 pKa = 3.42INSVNAEE288 pKa = 4.28FGNKK292 pKa = 7.03WMRR295 pKa = 11.84EE296 pKa = 3.4ASLRR300 pKa = 11.84MNKK303 pKa = 10.02LEE305 pKa = 4.32SLLDD309 pKa = 3.57KK310 pKa = 11.24LGDD313 pKa = 4.01GGSTVYY319 pKa = 10.32RR320 pKa = 11.84GRR322 pKa = 11.84LLEE325 pKa = 4.32ISSHH329 pKa = 5.11PLRR332 pKa = 11.84YY333 pKa = 9.47SLKK336 pKa = 8.63PLKK339 pKa = 10.23EE340 pKa = 3.8EE341 pKa = 5.21LMNQFWRR348 pKa = 11.84VMRR351 pKa = 11.84GEE353 pKa = 4.1EE354 pKa = 4.05VPIFSSQLQTDD365 pKa = 3.22IDD367 pKa = 4.22LLIFTLTEE375 pKa = 4.08GKK377 pKa = 9.91SFRR380 pKa = 11.84TWRR383 pKa = 11.84NSKK386 pKa = 7.86TSKK389 pKa = 10.14RR390 pKa = 11.84KK391 pKa = 8.1HH392 pKa = 5.21ACMLIKK398 pKa = 10.35KK399 pKa = 8.04VLKK402 pKa = 9.81EE403 pKa = 3.91SHH405 pKa = 6.03RR406 pKa = 11.84VAAAMKK412 pKa = 8.36EE413 pKa = 4.09LPTTSMSWEE422 pKa = 4.37DD423 pKa = 3.46YY424 pKa = 10.98AKK426 pKa = 10.51QFEE429 pKa = 4.31
MM1 pKa = 7.8KK2 pKa = 9.57MFKK5 pKa = 10.59YY6 pKa = 10.01MRR8 pKa = 11.84GDD10 pKa = 3.56ATWDD14 pKa = 3.1QEE16 pKa = 4.13KK17 pKa = 9.88QVRR20 pKa = 11.84RR21 pKa = 11.84ICKK24 pKa = 6.41WTEE27 pKa = 3.32RR28 pKa = 11.84GMKK31 pKa = 9.99LYY33 pKa = 11.02SFDD36 pKa = 3.35LTAATDD42 pKa = 3.74RR43 pKa = 11.84WPAWHH48 pKa = 6.37QKK50 pKa = 10.33LVVDD54 pKa = 4.28RR55 pKa = 11.84VFNKK59 pKa = 10.8GIGEE63 pKa = 3.87MWFRR67 pKa = 11.84LMTTTTPYY75 pKa = 9.35ATEE78 pKa = 4.05LKK80 pKa = 10.28RR81 pKa = 11.84YY82 pKa = 8.36IRR84 pKa = 11.84YY85 pKa = 9.86AIGQPMGAYY94 pKa = 10.19SSWAVLNMTHH104 pKa = 6.87HH105 pKa = 5.5YY106 pKa = 10.15VIRR109 pKa = 11.84LCAKK113 pKa = 10.16RR114 pKa = 11.84IGVKK118 pKa = 10.16ALYY121 pKa = 10.13SVLGDD126 pKa = 3.51DD127 pKa = 3.85VVIVGKK133 pKa = 10.11EE134 pKa = 3.5LADD137 pKa = 3.85EE138 pKa = 4.1YY139 pKa = 10.96RR140 pKa = 11.84SYY142 pKa = 9.78ITRR145 pKa = 11.84LGVKK149 pKa = 10.26LNLNKK154 pKa = 10.56SILNTDD160 pKa = 4.88FGPCSAEE167 pKa = 3.9FARR170 pKa = 11.84HH171 pKa = 4.76IVRR174 pKa = 11.84DD175 pKa = 3.47GHH177 pKa = 6.84IVGTFSPSLVRR188 pKa = 11.84NIYY191 pKa = 10.53SYY193 pKa = 10.33TDD195 pKa = 3.19VYY197 pKa = 11.69ASIDD201 pKa = 3.63LLRR204 pKa = 11.84EE205 pKa = 4.12VQSKK209 pKa = 10.08LGQVIHH215 pKa = 6.84VYY217 pKa = 10.81EE218 pKa = 4.39NTTLLPIPIVRR229 pKa = 11.84LLKK232 pKa = 9.98TFSFKK237 pKa = 10.93DD238 pKa = 3.33DD239 pKa = 3.61LLAIVTVPLTALKK252 pKa = 10.38LPRR255 pKa = 11.84VYY257 pKa = 10.47HH258 pKa = 5.59PEE260 pKa = 3.92KK261 pKa = 10.9FEE263 pKa = 4.25GDD265 pKa = 3.51PMVGYY270 pKa = 10.73DD271 pKa = 3.52KK272 pKa = 10.83FANPWYY278 pKa = 9.47EE279 pKa = 3.73CDD281 pKa = 3.42INSVNAEE288 pKa = 4.28FGNKK292 pKa = 7.03WMRR295 pKa = 11.84EE296 pKa = 3.4ASLRR300 pKa = 11.84MNKK303 pKa = 10.02LEE305 pKa = 4.32SLLDD309 pKa = 3.57KK310 pKa = 11.24LGDD313 pKa = 4.01GGSTVYY319 pKa = 10.32RR320 pKa = 11.84GRR322 pKa = 11.84LLEE325 pKa = 4.32ISSHH329 pKa = 5.11PLRR332 pKa = 11.84YY333 pKa = 9.47SLKK336 pKa = 8.63PLKK339 pKa = 10.23EE340 pKa = 3.8EE341 pKa = 5.21LMNQFWRR348 pKa = 11.84VMRR351 pKa = 11.84GEE353 pKa = 4.1EE354 pKa = 4.05VPIFSSQLQTDD365 pKa = 3.22IDD367 pKa = 4.22LLIFTLTEE375 pKa = 4.08GKK377 pKa = 9.91SFRR380 pKa = 11.84TWRR383 pKa = 11.84NSKK386 pKa = 7.86TSKK389 pKa = 10.14RR390 pKa = 11.84KK391 pKa = 8.1HH392 pKa = 5.21ACMLIKK398 pKa = 10.35KK399 pKa = 8.04VLKK402 pKa = 9.81EE403 pKa = 3.91SHH405 pKa = 6.03RR406 pKa = 11.84VAAAMKK412 pKa = 8.36EE413 pKa = 4.09LPTTSMSWEE422 pKa = 4.37DD423 pKa = 3.46YY424 pKa = 10.98AKK426 pKa = 10.51QFEE429 pKa = 4.31
Molecular weight: 49.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
429 |
429 |
429 |
429.0 |
49.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.594 ± 0.0 | 1.166 ± 0.0 |
5.128 ± 0.0 | 5.828 ± 0.0 |
3.963 ± 0.0 | 5.361 ± 0.0 |
2.331 ± 0.0 | 5.361 ± 0.0 |
7.692 ± 0.0 | 10.49 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.73 ± 0.0 | 3.263 ± 0.0 |
3.73 ± 0.0 | 2.331 ± 0.0 |
6.993 ± 0.0 | 6.527 ± 0.0 |
6.294 ± 0.0 | 6.993 ± 0.0 |
2.564 ± 0.0 | 4.662 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
