
Human PoSCV5-like circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
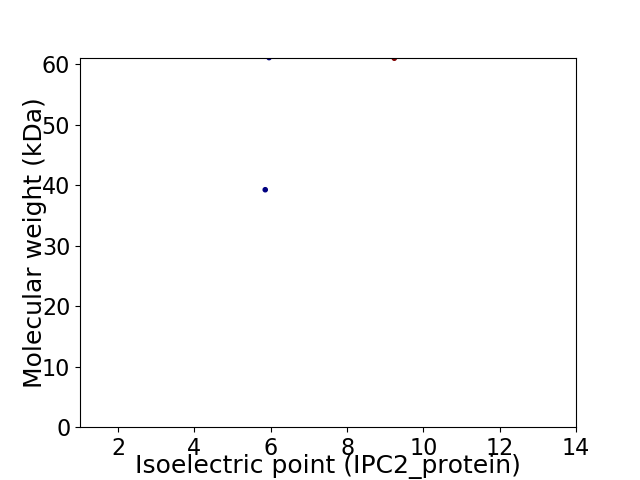
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
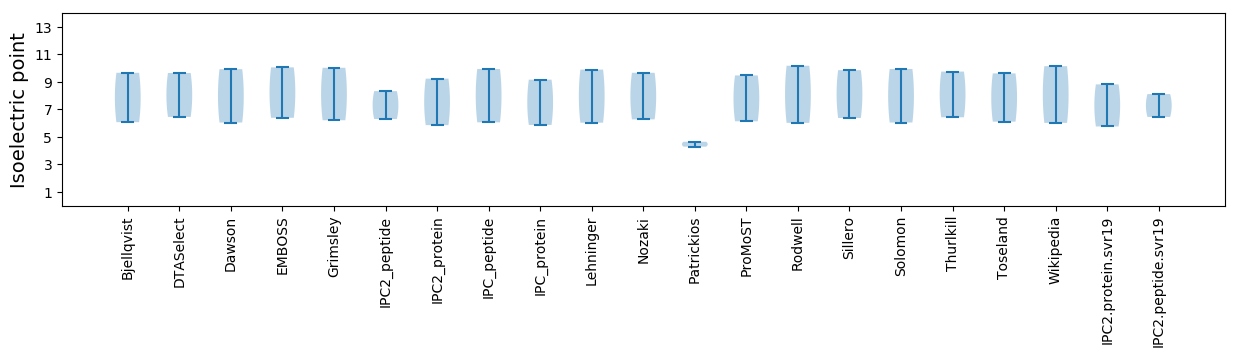
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D0WHQ3|A0A2D0WHQ3_9CIRC Cap protein OS=Human PoSCV5-like circular virus OX=1965018 PE=4 SV=1
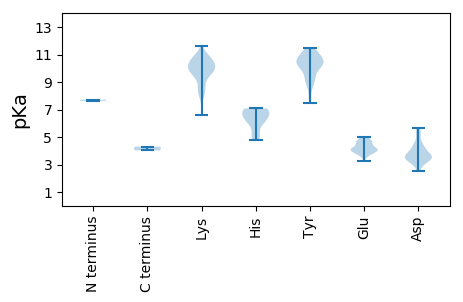
MM1 pKa = 7.63LSHH4 pKa = 7.08SSCKK8 pKa = 10.22LLNAKK13 pKa = 10.05RR14 pKa = 11.84FLLTYY19 pKa = 10.35NDD21 pKa = 3.46VXGKK25 pKa = 10.14IPSALDD31 pKa = 3.56LSEE34 pKa = 5.0HH35 pKa = 6.96LINLTSEE42 pKa = 3.63KK43 pKa = 9.96MEE45 pKa = 3.73FVVVSEE51 pKa = 4.38EE52 pKa = 4.14TAPTTGMIHH61 pKa = 5.05YY62 pKa = 8.3HH63 pKa = 6.2ALVILKK69 pKa = 10.46DD70 pKa = 3.53NLYY73 pKa = 10.69SRR75 pKa = 11.84KK76 pKa = 9.53LDD78 pKa = 3.52YY79 pKa = 10.78FDD81 pKa = 5.38YY82 pKa = 11.5LMIHH86 pKa = 6.61PNIEE90 pKa = 3.98AVRR93 pKa = 11.84GNAQKK98 pKa = 10.68AYY100 pKa = 9.98EE101 pKa = 3.97YY102 pKa = 10.59VIKK105 pKa = 10.46DD106 pKa = 3.3GKK108 pKa = 9.93YY109 pKa = 7.86WQTDD113 pKa = 3.32LNYY116 pKa = 10.39FKK118 pKa = 11.3VDD120 pKa = 3.16GGKK123 pKa = 10.36KK124 pKa = 8.8NNEE127 pKa = 3.23RR128 pKa = 11.84RR129 pKa = 11.84EE130 pKa = 4.07RR131 pKa = 11.84NKK133 pKa = 10.94LLMNGDD139 pKa = 3.05IFEE142 pKa = 4.67EE143 pKa = 4.56YY144 pKa = 10.22EE145 pKa = 4.14KK146 pKa = 11.59GNIGPIDD153 pKa = 5.08LIRR156 pKa = 11.84AAKK159 pKa = 9.44IRR161 pKa = 11.84AIKK164 pKa = 9.01VQYY167 pKa = 9.02NNNMTKK173 pKa = 9.26RR174 pKa = 11.84TPPLIMWFYY183 pKa = 11.24GNTGSGKK190 pKa = 6.62TRR192 pKa = 11.84KK193 pKa = 9.4AVEE196 pKa = 3.51IAEE199 pKa = 4.13EE200 pKa = 4.0LKK202 pKa = 11.0ARR204 pKa = 11.84IWISNDD210 pKa = 2.56NLKK213 pKa = 10.3WFDD216 pKa = 4.03GYY218 pKa = 10.87CGQEE222 pKa = 3.75VAVFDD227 pKa = 4.51DD228 pKa = 4.06LRR230 pKa = 11.84RR231 pKa = 11.84DD232 pKa = 3.86ILPTWAFLLRR242 pKa = 11.84LLDD245 pKa = 4.4RR246 pKa = 11.84YY247 pKa = 10.06PLIVQIKK254 pKa = 8.35GGFVKK259 pKa = 10.21WEE261 pKa = 3.87PKK263 pKa = 10.09IIIITCPVPPEE274 pKa = 4.33EE275 pKa = 4.74CFKK278 pKa = 10.48WYY280 pKa = 10.59NKK282 pKa = 9.82QGEE285 pKa = 4.28EE286 pKa = 4.08QQWDD290 pKa = 3.59RR291 pKa = 11.84QEE293 pKa = 3.9QLNRR297 pKa = 11.84RR298 pKa = 11.84LTWEE302 pKa = 4.12EE303 pKa = 3.88KK304 pKa = 8.11EE305 pKa = 4.08QVYY308 pKa = 9.0EE309 pKa = 4.45FPLSLGDD316 pKa = 3.84GEE318 pKa = 4.82EE319 pKa = 4.16MEE321 pKa = 4.64LTIEE325 pKa = 3.9NWKK328 pKa = 8.49TKK330 pKa = 10.16QADD333 pKa = 3.65DD334 pKa = 3.57NN335 pKa = 4.31
MM1 pKa = 7.63LSHH4 pKa = 7.08SSCKK8 pKa = 10.22LLNAKK13 pKa = 10.05RR14 pKa = 11.84FLLTYY19 pKa = 10.35NDD21 pKa = 3.46VXGKK25 pKa = 10.14IPSALDD31 pKa = 3.56LSEE34 pKa = 5.0HH35 pKa = 6.96LINLTSEE42 pKa = 3.63KK43 pKa = 9.96MEE45 pKa = 3.73FVVVSEE51 pKa = 4.38EE52 pKa = 4.14TAPTTGMIHH61 pKa = 5.05YY62 pKa = 8.3HH63 pKa = 6.2ALVILKK69 pKa = 10.46DD70 pKa = 3.53NLYY73 pKa = 10.69SRR75 pKa = 11.84KK76 pKa = 9.53LDD78 pKa = 3.52YY79 pKa = 10.78FDD81 pKa = 5.38YY82 pKa = 11.5LMIHH86 pKa = 6.61PNIEE90 pKa = 3.98AVRR93 pKa = 11.84GNAQKK98 pKa = 10.68AYY100 pKa = 9.98EE101 pKa = 3.97YY102 pKa = 10.59VIKK105 pKa = 10.46DD106 pKa = 3.3GKK108 pKa = 9.93YY109 pKa = 7.86WQTDD113 pKa = 3.32LNYY116 pKa = 10.39FKK118 pKa = 11.3VDD120 pKa = 3.16GGKK123 pKa = 10.36KK124 pKa = 8.8NNEE127 pKa = 3.23RR128 pKa = 11.84RR129 pKa = 11.84EE130 pKa = 4.07RR131 pKa = 11.84NKK133 pKa = 10.94LLMNGDD139 pKa = 3.05IFEE142 pKa = 4.67EE143 pKa = 4.56YY144 pKa = 10.22EE145 pKa = 4.14KK146 pKa = 11.59GNIGPIDD153 pKa = 5.08LIRR156 pKa = 11.84AAKK159 pKa = 9.44IRR161 pKa = 11.84AIKK164 pKa = 9.01VQYY167 pKa = 9.02NNNMTKK173 pKa = 9.26RR174 pKa = 11.84TPPLIMWFYY183 pKa = 11.24GNTGSGKK190 pKa = 6.62TRR192 pKa = 11.84KK193 pKa = 9.4AVEE196 pKa = 3.51IAEE199 pKa = 4.13EE200 pKa = 4.0LKK202 pKa = 11.0ARR204 pKa = 11.84IWISNDD210 pKa = 2.56NLKK213 pKa = 10.3WFDD216 pKa = 4.03GYY218 pKa = 10.87CGQEE222 pKa = 3.75VAVFDD227 pKa = 4.51DD228 pKa = 4.06LRR230 pKa = 11.84RR231 pKa = 11.84DD232 pKa = 3.86ILPTWAFLLRR242 pKa = 11.84LLDD245 pKa = 4.4RR246 pKa = 11.84YY247 pKa = 10.06PLIVQIKK254 pKa = 8.35GGFVKK259 pKa = 10.21WEE261 pKa = 3.87PKK263 pKa = 10.09IIIITCPVPPEE274 pKa = 4.33EE275 pKa = 4.74CFKK278 pKa = 10.48WYY280 pKa = 10.59NKK282 pKa = 9.82QGEE285 pKa = 4.28EE286 pKa = 4.08QQWDD290 pKa = 3.59RR291 pKa = 11.84QEE293 pKa = 3.9QLNRR297 pKa = 11.84RR298 pKa = 11.84LTWEE302 pKa = 4.12EE303 pKa = 3.88KK304 pKa = 8.11EE305 pKa = 4.08QVYY308 pKa = 9.0EE309 pKa = 4.45FPLSLGDD316 pKa = 3.84GEE318 pKa = 4.82EE319 pKa = 4.16MEE321 pKa = 4.64LTIEE325 pKa = 3.9NWKK328 pKa = 8.49TKK330 pKa = 10.16QADD333 pKa = 3.65DD334 pKa = 3.57NN335 pKa = 4.31
Molecular weight: 39.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D0WHQ3|A0A2D0WHQ3_9CIRC Cap protein OS=Human PoSCV5-like circular virus OX=1965018 PE=4 SV=1
MM1 pKa = 7.72AKK3 pKa = 9.97RR4 pKa = 11.84YY5 pKa = 9.36ASRR8 pKa = 11.84KK9 pKa = 7.79RR10 pKa = 11.84VYY12 pKa = 9.64RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.77ARR17 pKa = 11.84KK18 pKa = 7.99PIKK21 pKa = 9.65RR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 8.73RR27 pKa = 11.84AKK29 pKa = 10.62ARR31 pKa = 11.84STAKK35 pKa = 10.11RR36 pKa = 11.84YY37 pKa = 9.18FRR39 pKa = 11.84LKK41 pKa = 9.99RR42 pKa = 11.84AARR45 pKa = 11.84NYY47 pKa = 10.86SKK49 pKa = 10.84VKK51 pKa = 9.83KK52 pKa = 9.68IRR54 pKa = 11.84RR55 pKa = 11.84AKK57 pKa = 9.77RR58 pKa = 11.84QYY60 pKa = 9.43KK61 pKa = 8.42TISGKK66 pKa = 8.26GVKK69 pKa = 9.78NKK71 pKa = 10.11LSKK74 pKa = 10.87LSISEE79 pKa = 4.8FIQLPYY85 pKa = 10.56YY86 pKa = 10.6DD87 pKa = 5.67LIRR90 pKa = 11.84LASMVCTRR98 pKa = 11.84HH99 pKa = 6.22SPTGYY104 pKa = 9.68PLTEE108 pKa = 3.69QAMQSFANLYY118 pKa = 9.91SEE120 pKa = 4.3QYY122 pKa = 10.16LYY124 pKa = 11.34KK125 pKa = 10.61LGAILPEE132 pKa = 3.96KK133 pKa = 10.62LFVNSNPFIIGADD146 pKa = 3.31RR147 pKa = 11.84SFTPYY152 pKa = 10.9LFGFNPILTPVTCGQKK168 pKa = 8.28MQIYY172 pKa = 9.9ASLYY176 pKa = 10.19SKK178 pKa = 10.47FKK180 pKa = 9.54YY181 pKa = 10.48VGVKK185 pKa = 9.95VSWHH189 pKa = 5.24PRR191 pKa = 11.84NRR193 pKa = 11.84AVSSYY198 pKa = 10.55LPSEE202 pKa = 4.44FKK204 pKa = 10.87VKK206 pKa = 10.56YY207 pKa = 10.37DD208 pKa = 3.59SDD210 pKa = 4.35VLSMAALTPNPAEE223 pKa = 4.86DD224 pKa = 4.32GVPSTIAQGNAMFTPKK240 pKa = 10.4DD241 pKa = 3.53RR242 pKa = 11.84LNQTIEE248 pKa = 4.04MTRR251 pKa = 11.84SEE253 pKa = 4.59ANVSGNYY260 pKa = 9.25YY261 pKa = 7.49MHH263 pKa = 7.08VYY265 pKa = 10.16FGKK268 pKa = 10.38QIEE271 pKa = 4.52SSEE274 pKa = 4.26TWDD277 pKa = 3.32LPYY280 pKa = 10.8YY281 pKa = 10.33RR282 pKa = 11.84WRR284 pKa = 11.84QMSGGQKK291 pKa = 9.72EE292 pKa = 4.81LSLQHH297 pKa = 6.37EE298 pKa = 4.59KK299 pKa = 10.9AKK301 pKa = 10.74HH302 pKa = 4.8NNKK305 pKa = 9.38GRR307 pKa = 11.84VKK309 pKa = 10.83ASLFYY314 pKa = 10.53DD315 pKa = 3.43IQTLLPCIEE324 pKa = 4.53TNSKK328 pKa = 8.82LHH330 pKa = 6.11KK331 pKa = 10.18VYY333 pKa = 11.13DD334 pKa = 3.55MSKK337 pKa = 8.64PFSFFVRR344 pKa = 11.84PMVVDD349 pKa = 3.81SSTVKK354 pKa = 10.43EE355 pKa = 4.13PNEE358 pKa = 3.91RR359 pKa = 11.84GAEE362 pKa = 4.14NIDD365 pKa = 4.27TIDD368 pKa = 4.16CNCQHH373 pKa = 6.79EE374 pKa = 4.87SLTKK378 pKa = 9.66GLKK381 pKa = 9.22HH382 pKa = 6.22LGYY385 pKa = 10.47RR386 pKa = 11.84SFDD389 pKa = 3.43HH390 pKa = 7.04ALVPSTIEE398 pKa = 3.75IPQDD402 pKa = 3.45NQEE405 pKa = 4.72LSDD408 pKa = 3.64LTQDD412 pKa = 2.56RR413 pKa = 11.84WYY415 pKa = 11.27YY416 pKa = 11.28NSDD419 pKa = 3.2DD420 pKa = 5.15DD421 pKa = 5.56NFFNPTLFWYY431 pKa = 10.55CFTTSDD437 pKa = 3.76NPVDD441 pKa = 3.69ASYY444 pKa = 11.05KK445 pKa = 9.78SHH447 pKa = 7.0SPYY450 pKa = 10.08QALASSFNNAQVPAGAPNGNARR472 pKa = 11.84YY473 pKa = 8.93EE474 pKa = 4.17LSSHH478 pKa = 6.81FGTYY482 pKa = 10.15NDD484 pKa = 4.51ALNQYY489 pKa = 10.36GYY491 pKa = 11.18FDD493 pKa = 3.34VTFYY497 pKa = 10.81TKK499 pKa = 9.92WKK501 pKa = 9.94GEE503 pKa = 3.91RR504 pKa = 11.84PLVSKK509 pKa = 10.74SSYY512 pKa = 9.05TGIVPSEE519 pKa = 4.11TGVTGLSTIISNN531 pKa = 4.03
MM1 pKa = 7.72AKK3 pKa = 9.97RR4 pKa = 11.84YY5 pKa = 9.36ASRR8 pKa = 11.84KK9 pKa = 7.79RR10 pKa = 11.84VYY12 pKa = 9.64RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.77ARR17 pKa = 11.84KK18 pKa = 7.99PIKK21 pKa = 9.65RR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 8.73RR27 pKa = 11.84AKK29 pKa = 10.62ARR31 pKa = 11.84STAKK35 pKa = 10.11RR36 pKa = 11.84YY37 pKa = 9.18FRR39 pKa = 11.84LKK41 pKa = 9.99RR42 pKa = 11.84AARR45 pKa = 11.84NYY47 pKa = 10.86SKK49 pKa = 10.84VKK51 pKa = 9.83KK52 pKa = 9.68IRR54 pKa = 11.84RR55 pKa = 11.84AKK57 pKa = 9.77RR58 pKa = 11.84QYY60 pKa = 9.43KK61 pKa = 8.42TISGKK66 pKa = 8.26GVKK69 pKa = 9.78NKK71 pKa = 10.11LSKK74 pKa = 10.87LSISEE79 pKa = 4.8FIQLPYY85 pKa = 10.56YY86 pKa = 10.6DD87 pKa = 5.67LIRR90 pKa = 11.84LASMVCTRR98 pKa = 11.84HH99 pKa = 6.22SPTGYY104 pKa = 9.68PLTEE108 pKa = 3.69QAMQSFANLYY118 pKa = 9.91SEE120 pKa = 4.3QYY122 pKa = 10.16LYY124 pKa = 11.34KK125 pKa = 10.61LGAILPEE132 pKa = 3.96KK133 pKa = 10.62LFVNSNPFIIGADD146 pKa = 3.31RR147 pKa = 11.84SFTPYY152 pKa = 10.9LFGFNPILTPVTCGQKK168 pKa = 8.28MQIYY172 pKa = 9.9ASLYY176 pKa = 10.19SKK178 pKa = 10.47FKK180 pKa = 9.54YY181 pKa = 10.48VGVKK185 pKa = 9.95VSWHH189 pKa = 5.24PRR191 pKa = 11.84NRR193 pKa = 11.84AVSSYY198 pKa = 10.55LPSEE202 pKa = 4.44FKK204 pKa = 10.87VKK206 pKa = 10.56YY207 pKa = 10.37DD208 pKa = 3.59SDD210 pKa = 4.35VLSMAALTPNPAEE223 pKa = 4.86DD224 pKa = 4.32GVPSTIAQGNAMFTPKK240 pKa = 10.4DD241 pKa = 3.53RR242 pKa = 11.84LNQTIEE248 pKa = 4.04MTRR251 pKa = 11.84SEE253 pKa = 4.59ANVSGNYY260 pKa = 9.25YY261 pKa = 7.49MHH263 pKa = 7.08VYY265 pKa = 10.16FGKK268 pKa = 10.38QIEE271 pKa = 4.52SSEE274 pKa = 4.26TWDD277 pKa = 3.32LPYY280 pKa = 10.8YY281 pKa = 10.33RR282 pKa = 11.84WRR284 pKa = 11.84QMSGGQKK291 pKa = 9.72EE292 pKa = 4.81LSLQHH297 pKa = 6.37EE298 pKa = 4.59KK299 pKa = 10.9AKK301 pKa = 10.74HH302 pKa = 4.8NNKK305 pKa = 9.38GRR307 pKa = 11.84VKK309 pKa = 10.83ASLFYY314 pKa = 10.53DD315 pKa = 3.43IQTLLPCIEE324 pKa = 4.53TNSKK328 pKa = 8.82LHH330 pKa = 6.11KK331 pKa = 10.18VYY333 pKa = 11.13DD334 pKa = 3.55MSKK337 pKa = 8.64PFSFFVRR344 pKa = 11.84PMVVDD349 pKa = 3.81SSTVKK354 pKa = 10.43EE355 pKa = 4.13PNEE358 pKa = 3.91RR359 pKa = 11.84GAEE362 pKa = 4.14NIDD365 pKa = 4.27TIDD368 pKa = 4.16CNCQHH373 pKa = 6.79EE374 pKa = 4.87SLTKK378 pKa = 9.66GLKK381 pKa = 9.22HH382 pKa = 6.22LGYY385 pKa = 10.47RR386 pKa = 11.84SFDD389 pKa = 3.43HH390 pKa = 7.04ALVPSTIEE398 pKa = 3.75IPQDD402 pKa = 3.45NQEE405 pKa = 4.72LSDD408 pKa = 3.64LTQDD412 pKa = 2.56RR413 pKa = 11.84WYY415 pKa = 11.27YY416 pKa = 11.28NSDD419 pKa = 3.2DD420 pKa = 5.15DD421 pKa = 5.56NFFNPTLFWYY431 pKa = 10.55CFTTSDD437 pKa = 3.76NPVDD441 pKa = 3.69ASYY444 pKa = 11.05KK445 pKa = 9.78SHH447 pKa = 7.0SPYY450 pKa = 10.08QALASSFNNAQVPAGAPNGNARR472 pKa = 11.84YY473 pKa = 8.93EE474 pKa = 4.17LSSHH478 pKa = 6.81FGTYY482 pKa = 10.15NDD484 pKa = 4.51ALNQYY489 pKa = 10.36GYY491 pKa = 11.18FDD493 pKa = 3.34VTFYY497 pKa = 10.81TKK499 pKa = 9.92WKK501 pKa = 9.94GEE503 pKa = 3.91RR504 pKa = 11.84PLVSKK509 pKa = 10.74SSYY512 pKa = 9.05TGIVPSEE519 pKa = 4.11TGVTGLSTIISNN531 pKa = 4.03
Molecular weight: 61.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
866 |
335 |
531 |
433.0 |
50.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.774 ± 0.611 | 1.155 ± 0.024 |
4.965 ± 0.615 | 6.005 ± 1.806 |
4.388 ± 0.493 | 5.196 ± 0.291 |
1.848 ± 0.217 | 5.543 ± 1.175 |
8.199 ± 0.28 | 8.199 ± 1.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.194 ± 0.119 | 6.005 ± 0.344 |
4.85 ± 0.593 | 3.811 ± 0.14 |
6.005 ± 0.386 | 7.39 ± 2.513 |
5.312 ± 0.511 | 5.081 ± 0.186 |
1.848 ± 0.696 | 6.12 ± 0.823 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
