
Corynebacterium phage EmiRose
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
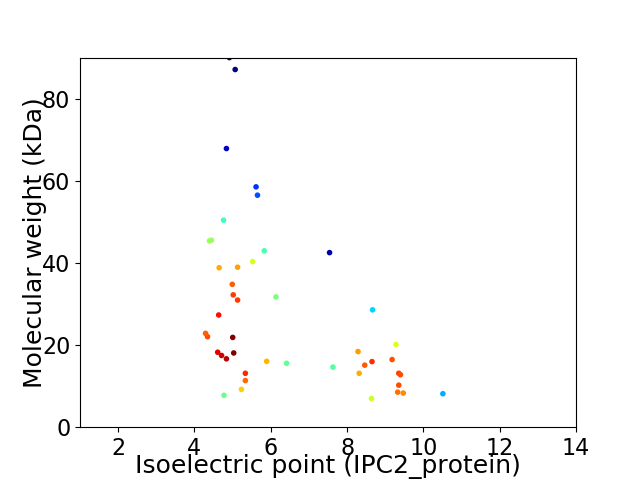
Virtual 2D-PAGE plot for 46 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A649VNX4|A0A649VNX4_9CAUD Portal protein OS=Corynebacterium phage EmiRose OX=2565372 GN=3 PE=4 SV=1
MM1 pKa = 7.94DD2 pKa = 3.45MRR4 pKa = 11.84YY5 pKa = 6.87TTMPVDD11 pKa = 3.9GVTLASYY18 pKa = 10.95SKK20 pKa = 11.09ALLPRR25 pKa = 11.84PQHH28 pKa = 6.03TGDD31 pKa = 3.33AGIDD35 pKa = 3.71LRR37 pKa = 11.84CASAFYY43 pKa = 10.71LEE45 pKa = 5.12PGEE48 pKa = 4.19TMQIGTGVVVDD59 pKa = 4.47NIDD62 pKa = 3.51RR63 pKa = 11.84YY64 pKa = 10.07IVGMVCSRR72 pKa = 11.84SGLAAHH78 pKa = 7.29DD79 pKa = 4.18GVHH82 pKa = 5.77VLNAPGIIDD91 pKa = 3.24SMYY94 pKa = 9.41RR95 pKa = 11.84GEE97 pKa = 4.22IAVILHH103 pKa = 6.0NASSHH108 pKa = 5.54RR109 pKa = 11.84CVVEE113 pKa = 3.8AGRR116 pKa = 11.84RR117 pKa = 11.84IAQLVFTPVCPTVAPVQDD135 pKa = 3.57TDD137 pKa = 3.58RR138 pKa = 11.84GEE140 pKa = 4.43GGLGSTDD147 pKa = 3.4SINPRR152 pKa = 11.84EE153 pKa = 4.26YY154 pKa = 10.74AVPQHH159 pKa = 5.75YY160 pKa = 10.85VIGDD164 pKa = 3.58AFDD167 pKa = 3.75GHH169 pKa = 7.56DD170 pKa = 5.22DD171 pKa = 4.36GADD174 pKa = 3.33DD175 pKa = 4.73AANLFEE181 pKa = 7.11ADD183 pKa = 5.23DD184 pKa = 4.36EE185 pKa = 4.53LLSDD189 pKa = 4.46EE190 pKa = 4.8DD191 pKa = 5.35RR192 pKa = 11.84EE193 pKa = 4.6IADD196 pKa = 4.6LLLGDD201 pKa = 4.78GDD203 pKa = 6.16DD204 pKa = 4.53MLTDD208 pKa = 4.91DD209 pKa = 5.57LPSDD213 pKa = 3.81MDD215 pKa = 4.15DD216 pKa = 4.63LVDD219 pKa = 5.17KK220 pKa = 10.21VTTLIFKK227 pKa = 10.35AAAANLNDD235 pKa = 5.36DD236 pKa = 3.97GTIPDD241 pKa = 4.95DD242 pKa = 3.7NAGSDD247 pKa = 4.21DD248 pKa = 4.08VVSHH252 pKa = 6.88PKK254 pKa = 10.14HH255 pKa = 5.2YY256 pKa = 10.45AANRR260 pKa = 11.84DD261 pKa = 3.78FPGLEE266 pKa = 4.35AILFTRR272 pKa = 11.84TMRR275 pKa = 11.84FGHH278 pKa = 6.04GNAVKK283 pKa = 10.14YY284 pKa = 10.0LWRR287 pKa = 11.84HH288 pKa = 5.06AYY290 pKa = 9.38KK291 pKa = 10.57GRR293 pKa = 11.84PVEE296 pKa = 4.27DD297 pKa = 3.21LRR299 pKa = 11.84KK300 pKa = 10.03ARR302 pKa = 11.84WYY304 pKa = 10.17LVNSIEE310 pKa = 4.91GGSQLLHH317 pKa = 6.76LKK319 pKa = 9.63NAPDD323 pKa = 3.39NADD326 pKa = 3.52NEE328 pKa = 4.55RR329 pKa = 11.84EE330 pKa = 4.03LLDD333 pKa = 4.01RR334 pKa = 11.84MDD336 pKa = 3.9YY337 pKa = 9.66MNYY340 pKa = 9.68MYY342 pKa = 10.09IEE344 pKa = 4.96CIGGRR349 pKa = 11.84GNKK352 pKa = 9.45HH353 pKa = 5.47EE354 pKa = 4.31ALKK357 pKa = 11.01DD358 pKa = 3.7VAKK361 pKa = 10.64LLAYY365 pKa = 9.82RR366 pKa = 11.84VMVGLIFYY374 pKa = 10.05GEE376 pKa = 4.34EE377 pKa = 3.76YY378 pKa = 10.73AALLRR383 pKa = 11.84GHH385 pKa = 6.4SLLGGDD391 pKa = 4.02TEE393 pKa = 5.49AMSEE397 pKa = 4.18TQLIRR402 pKa = 11.84ILDD405 pKa = 3.94CAIDD409 pKa = 3.76VYY411 pKa = 9.15TTLEE415 pKa = 3.95EE416 pKa = 4.11
MM1 pKa = 7.94DD2 pKa = 3.45MRR4 pKa = 11.84YY5 pKa = 6.87TTMPVDD11 pKa = 3.9GVTLASYY18 pKa = 10.95SKK20 pKa = 11.09ALLPRR25 pKa = 11.84PQHH28 pKa = 6.03TGDD31 pKa = 3.33AGIDD35 pKa = 3.71LRR37 pKa = 11.84CASAFYY43 pKa = 10.71LEE45 pKa = 5.12PGEE48 pKa = 4.19TMQIGTGVVVDD59 pKa = 4.47NIDD62 pKa = 3.51RR63 pKa = 11.84YY64 pKa = 10.07IVGMVCSRR72 pKa = 11.84SGLAAHH78 pKa = 7.29DD79 pKa = 4.18GVHH82 pKa = 5.77VLNAPGIIDD91 pKa = 3.24SMYY94 pKa = 9.41RR95 pKa = 11.84GEE97 pKa = 4.22IAVILHH103 pKa = 6.0NASSHH108 pKa = 5.54RR109 pKa = 11.84CVVEE113 pKa = 3.8AGRR116 pKa = 11.84RR117 pKa = 11.84IAQLVFTPVCPTVAPVQDD135 pKa = 3.57TDD137 pKa = 3.58RR138 pKa = 11.84GEE140 pKa = 4.43GGLGSTDD147 pKa = 3.4SINPRR152 pKa = 11.84EE153 pKa = 4.26YY154 pKa = 10.74AVPQHH159 pKa = 5.75YY160 pKa = 10.85VIGDD164 pKa = 3.58AFDD167 pKa = 3.75GHH169 pKa = 7.56DD170 pKa = 5.22DD171 pKa = 4.36GADD174 pKa = 3.33DD175 pKa = 4.73AANLFEE181 pKa = 7.11ADD183 pKa = 5.23DD184 pKa = 4.36EE185 pKa = 4.53LLSDD189 pKa = 4.46EE190 pKa = 4.8DD191 pKa = 5.35RR192 pKa = 11.84EE193 pKa = 4.6IADD196 pKa = 4.6LLLGDD201 pKa = 4.78GDD203 pKa = 6.16DD204 pKa = 4.53MLTDD208 pKa = 4.91DD209 pKa = 5.57LPSDD213 pKa = 3.81MDD215 pKa = 4.15DD216 pKa = 4.63LVDD219 pKa = 5.17KK220 pKa = 10.21VTTLIFKK227 pKa = 10.35AAAANLNDD235 pKa = 5.36DD236 pKa = 3.97GTIPDD241 pKa = 4.95DD242 pKa = 3.7NAGSDD247 pKa = 4.21DD248 pKa = 4.08VVSHH252 pKa = 6.88PKK254 pKa = 10.14HH255 pKa = 5.2YY256 pKa = 10.45AANRR260 pKa = 11.84DD261 pKa = 3.78FPGLEE266 pKa = 4.35AILFTRR272 pKa = 11.84TMRR275 pKa = 11.84FGHH278 pKa = 6.04GNAVKK283 pKa = 10.14YY284 pKa = 10.0LWRR287 pKa = 11.84HH288 pKa = 5.06AYY290 pKa = 9.38KK291 pKa = 10.57GRR293 pKa = 11.84PVEE296 pKa = 4.27DD297 pKa = 3.21LRR299 pKa = 11.84KK300 pKa = 10.03ARR302 pKa = 11.84WYY304 pKa = 10.17LVNSIEE310 pKa = 4.91GGSQLLHH317 pKa = 6.76LKK319 pKa = 9.63NAPDD323 pKa = 3.39NADD326 pKa = 3.52NEE328 pKa = 4.55RR329 pKa = 11.84EE330 pKa = 4.03LLDD333 pKa = 4.01RR334 pKa = 11.84MDD336 pKa = 3.9YY337 pKa = 9.66MNYY340 pKa = 9.68MYY342 pKa = 10.09IEE344 pKa = 4.96CIGGRR349 pKa = 11.84GNKK352 pKa = 9.45HH353 pKa = 5.47EE354 pKa = 4.31ALKK357 pKa = 11.01DD358 pKa = 3.7VAKK361 pKa = 10.64LLAYY365 pKa = 9.82RR366 pKa = 11.84VMVGLIFYY374 pKa = 10.05GEE376 pKa = 4.34EE377 pKa = 3.76YY378 pKa = 10.73AALLRR383 pKa = 11.84GHH385 pKa = 6.4SLLGGDD391 pKa = 4.02TEE393 pKa = 5.49AMSEE397 pKa = 4.18TQLIRR402 pKa = 11.84ILDD405 pKa = 3.94CAIDD409 pKa = 3.76VYY411 pKa = 9.15TTLEE415 pKa = 3.95EE416 pKa = 4.11
Molecular weight: 45.66 kDa
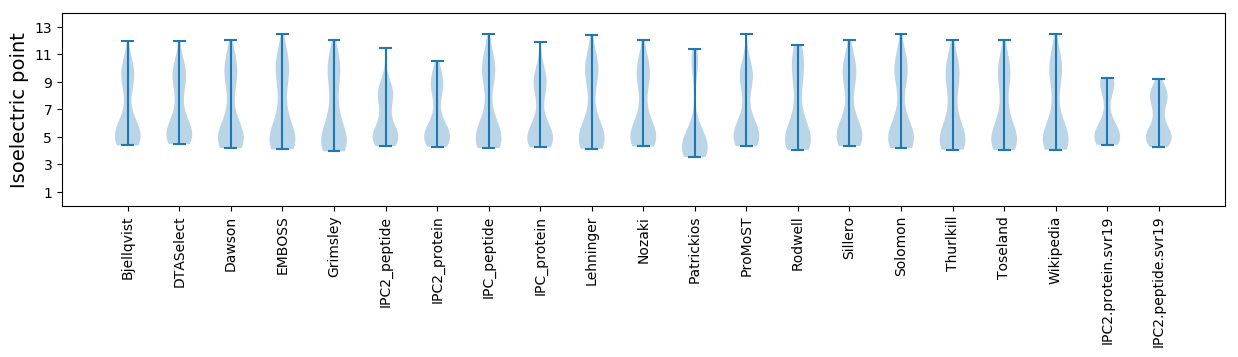
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A649VPG7|A0A649VPG7_9CAUD Head-to-tail stopper OS=Corynebacterium phage EmiRose OX=2565372 GN=9 PE=4 SV=1
MM1 pKa = 7.0GTNDD5 pKa = 3.11VPGPGFIRR13 pKa = 11.84SGDD16 pKa = 3.54RR17 pKa = 11.84KK18 pKa = 9.87NKK20 pKa = 9.57ARR22 pKa = 11.84ASSWAWQRR30 pKa = 11.84RR31 pKa = 11.84VKK33 pKa = 10.78AEE35 pKa = 3.72RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84LEE40 pKa = 4.19KK41 pKa = 10.84DD42 pKa = 2.92PSLAVCWICNEE53 pKa = 4.68PIDD56 pKa = 3.84MTLPYY61 pKa = 10.1QHH63 pKa = 6.8ARR65 pKa = 11.84AFTLDD70 pKa = 3.21HH71 pKa = 6.63LVPIGRR77 pKa = 11.84GGALDD82 pKa = 4.05GDD84 pKa = 4.09TRR86 pKa = 11.84PAHH89 pKa = 6.4LSCNAKK95 pKa = 10.4RR96 pKa = 11.84GDD98 pKa = 3.28GRR100 pKa = 11.84RR101 pKa = 11.84KK102 pKa = 10.01KK103 pKa = 10.53KK104 pKa = 9.62EE105 pKa = 4.15SKK107 pKa = 8.32PTTIIGWW114 pKa = 3.57
MM1 pKa = 7.0GTNDD5 pKa = 3.11VPGPGFIRR13 pKa = 11.84SGDD16 pKa = 3.54RR17 pKa = 11.84KK18 pKa = 9.87NKK20 pKa = 9.57ARR22 pKa = 11.84ASSWAWQRR30 pKa = 11.84RR31 pKa = 11.84VKK33 pKa = 10.78AEE35 pKa = 3.72RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84LEE40 pKa = 4.19KK41 pKa = 10.84DD42 pKa = 2.92PSLAVCWICNEE53 pKa = 4.68PIDD56 pKa = 3.84MTLPYY61 pKa = 10.1QHH63 pKa = 6.8ARR65 pKa = 11.84AFTLDD70 pKa = 3.21HH71 pKa = 6.63LVPIGRR77 pKa = 11.84GGALDD82 pKa = 4.05GDD84 pKa = 4.09TRR86 pKa = 11.84PAHH89 pKa = 6.4LSCNAKK95 pKa = 10.4RR96 pKa = 11.84GDD98 pKa = 3.28GRR100 pKa = 11.84RR101 pKa = 11.84KK102 pKa = 10.01KK103 pKa = 10.53KK104 pKa = 9.62EE105 pKa = 4.15SKK107 pKa = 8.32PTTIIGWW114 pKa = 3.57
Molecular weight: 12.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11873 |
60 |
886 |
258.1 |
27.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.657 ± 0.57 | 0.623 ± 0.097 |
6.73 ± 0.301 | 5.601 ± 0.329 |
2.577 ± 0.151 | 8.17 ± 0.431 |
1.583 ± 0.145 | 4.641 ± 0.221 |
4.54 ± 0.263 | 8.709 ± 0.271 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.173 ± 0.143 | 3.184 ± 0.214 |
5.55 ± 0.306 | 3.31 ± 0.212 |
5.938 ± 0.333 | 6.376 ± 0.328 |
6.831 ± 0.357 | 7.403 ± 0.24 |
1.752 ± 0.14 | 2.653 ± 0.229 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
