
Cognatiyoonia koreensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Cognatiyoonia
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
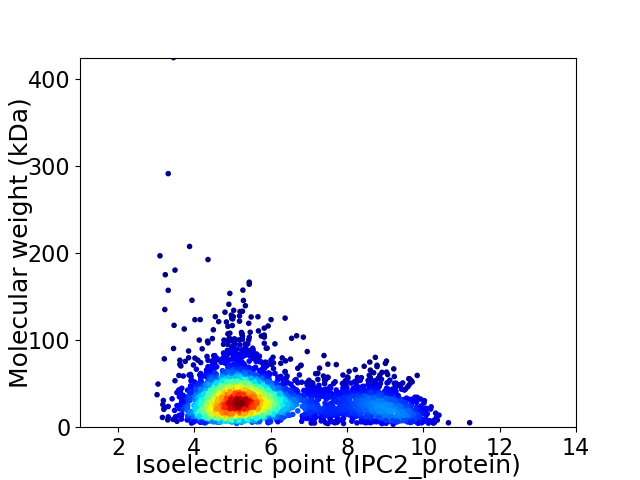
Virtual 2D-PAGE plot for 3582 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
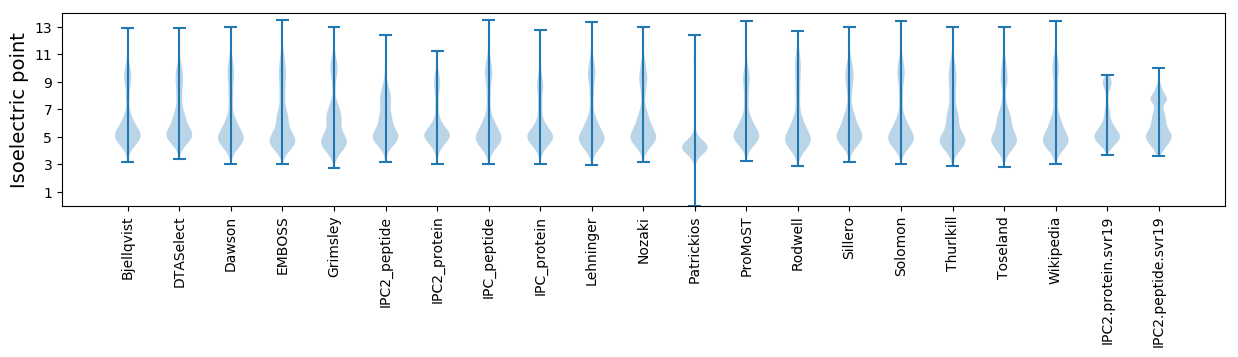
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0N387|A0A1I0N387_9RHOB Uncharacterized protein OS=Cognatiyoonia koreensis OX=364200 GN=SAMN04488515_0391 PE=4 SV=1
MM1 pKa = 7.19PTIDD5 pKa = 3.68VTDD8 pKa = 3.85IAANFTMSFGYY19 pKa = 10.21QFSSYY24 pKa = 10.02VGSGSTFYY32 pKa = 10.81SWEE35 pKa = 4.19TNGGHH40 pKa = 7.08TITLTGTGITTGVGNLPTGGTITGMLIDD68 pKa = 5.05LNGTAPSDD76 pKa = 3.51VDD78 pKa = 3.84VEE80 pKa = 4.11ISGIEE85 pKa = 3.97NKK87 pKa = 10.53NITDD91 pKa = 3.47IVFVAGPDD99 pKa = 3.65SEE101 pKa = 4.42NTLWATVLDD110 pKa = 4.01GDD112 pKa = 4.35VTVDD116 pKa = 3.9GAPTEE121 pKa = 4.1DD122 pKa = 3.86FRR124 pKa = 11.84VEE126 pKa = 3.74INTDD130 pKa = 3.57FDD132 pKa = 3.87EE133 pKa = 4.94VSFGIHH139 pKa = 5.98NGEE142 pKa = 4.46DD143 pKa = 2.98ATIRR147 pKa = 11.84GDD149 pKa = 3.87FFGFTYY155 pKa = 10.56FGGVQVLEE163 pKa = 4.61GTAHH167 pKa = 6.58FSAGEE172 pKa = 3.85TDD174 pKa = 4.59FEE176 pKa = 4.94GFIYY180 pKa = 9.97WFNPAQTVRR189 pKa = 11.84GNARR193 pKa = 11.84LLGGDD198 pKa = 3.95DD199 pKa = 3.86TIIWTEE205 pKa = 3.93DD206 pKa = 2.98TRR208 pKa = 11.84SYY210 pKa = 10.24PSSYY214 pKa = 10.61VYY216 pKa = 10.57AYY218 pKa = 10.45ADD220 pKa = 3.28ASFVSGSAVVVGGDD234 pKa = 3.76DD235 pKa = 5.03LLDD238 pKa = 3.75ARR240 pKa = 11.84NAGDD244 pKa = 4.13GIGAFLVGDD253 pKa = 4.66AYY255 pKa = 10.28HH256 pKa = 5.93TTAATAMVIGGDD268 pKa = 4.37DD269 pKa = 3.34ILFGANASDD278 pKa = 3.72GAPTNRR284 pKa = 11.84LYY286 pKa = 11.53GDD288 pKa = 3.58VVVNFTDD295 pKa = 3.52TLIAGDD301 pKa = 3.56DD302 pKa = 4.04TIYY305 pKa = 11.19GRR307 pKa = 11.84AGNDD311 pKa = 3.26EE312 pKa = 4.47IYY314 pKa = 10.88GDD316 pKa = 3.89TGSTGTNYY324 pKa = 9.94IGGDD328 pKa = 3.45DD329 pKa = 5.21LIYY332 pKa = 10.79GGSDD336 pKa = 3.9GEE338 pKa = 4.11DD339 pKa = 2.99LMYY342 pKa = 10.77GGRR345 pKa = 11.84GNDD348 pKa = 2.63RR349 pKa = 11.84MYY351 pKa = 10.69GGKK354 pKa = 10.46GIVEE358 pKa = 3.94MRR360 pKa = 11.84GDD362 pKa = 3.47QDD364 pKa = 3.49NDD366 pKa = 3.03KK367 pKa = 10.84LYY369 pKa = 11.04AGDD372 pKa = 4.04GTTTAYY378 pKa = 10.36GGSGDD383 pKa = 5.14DD384 pKa = 4.38LIKK387 pKa = 10.51VTGDD391 pKa = 3.59GEE393 pKa = 4.62GSFYY397 pKa = 10.84GGSGIDD403 pKa = 3.27RR404 pKa = 11.84ISYY407 pKa = 9.77YY408 pKa = 10.46DD409 pKa = 3.56AEE411 pKa = 4.63GGITIDD417 pKa = 4.51LGANTISRR425 pKa = 11.84YY426 pKa = 8.71WADD429 pKa = 4.94DD430 pKa = 3.37DD431 pKa = 4.82TISNFEE437 pKa = 4.15NVSGSKK443 pKa = 10.23VGNNKK448 pKa = 9.17IFGTSGANDD457 pKa = 3.28IDD459 pKa = 4.27TYY461 pKa = 11.48DD462 pKa = 4.35GNDD465 pKa = 3.06KK466 pKa = 11.11VYY468 pKa = 11.06GGGGGDD474 pKa = 4.29KK475 pKa = 10.53ISLGKK480 pKa = 10.88GNDD483 pKa = 3.36YY484 pKa = 11.24VKK486 pKa = 11.14ADD488 pKa = 4.64FGANTYY494 pKa = 9.7EE495 pKa = 4.44GSSGLDD501 pKa = 3.03TISYY505 pKa = 9.69YY506 pKa = 11.05NSNDD510 pKa = 3.11GVIVDD515 pKa = 4.74LKK517 pKa = 10.96HH518 pKa = 6.39NEE520 pKa = 4.0AEE522 pKa = 4.17GGYY525 pKa = 10.79AEE527 pKa = 5.58GDD529 pKa = 3.7TISGFEE535 pKa = 4.19RR536 pKa = 11.84LTGSTEE542 pKa = 3.77DD543 pKa = 3.63DD544 pKa = 3.49KK545 pKa = 11.89LYY547 pKa = 11.16GSDD550 pKa = 4.12GGNILKK556 pKa = 10.52GGSGEE561 pKa = 3.99DD562 pKa = 3.8SLWGRR567 pKa = 11.84DD568 pKa = 4.05GNDD571 pKa = 3.07KK572 pKa = 11.09LYY574 pKa = 11.13GGSNSDD580 pKa = 3.46RR581 pKa = 11.84FDD583 pKa = 3.63GGEE586 pKa = 4.38GNDD589 pKa = 3.7KK590 pKa = 10.91LYY592 pKa = 11.14GGSGVDD598 pKa = 3.11TFHH601 pKa = 7.45FDD603 pKa = 4.17KK604 pKa = 11.3GDD606 pKa = 3.95DD607 pKa = 3.8FDD609 pKa = 4.83TVMDD613 pKa = 4.23FQNNADD619 pKa = 4.43RR620 pKa = 11.84IEE622 pKa = 4.45IDD624 pKa = 4.06GYY626 pKa = 11.52SSGFDD631 pKa = 2.99AFDD634 pKa = 3.57YY635 pKa = 8.49ATQVGTDD642 pKa = 3.2VVFNFGGGDD651 pKa = 3.58GLIVEE656 pKa = 4.81NVLIGQLGNDD666 pKa = 3.88LFVVV670 pKa = 4.15
MM1 pKa = 7.19PTIDD5 pKa = 3.68VTDD8 pKa = 3.85IAANFTMSFGYY19 pKa = 10.21QFSSYY24 pKa = 10.02VGSGSTFYY32 pKa = 10.81SWEE35 pKa = 4.19TNGGHH40 pKa = 7.08TITLTGTGITTGVGNLPTGGTITGMLIDD68 pKa = 5.05LNGTAPSDD76 pKa = 3.51VDD78 pKa = 3.84VEE80 pKa = 4.11ISGIEE85 pKa = 3.97NKK87 pKa = 10.53NITDD91 pKa = 3.47IVFVAGPDD99 pKa = 3.65SEE101 pKa = 4.42NTLWATVLDD110 pKa = 4.01GDD112 pKa = 4.35VTVDD116 pKa = 3.9GAPTEE121 pKa = 4.1DD122 pKa = 3.86FRR124 pKa = 11.84VEE126 pKa = 3.74INTDD130 pKa = 3.57FDD132 pKa = 3.87EE133 pKa = 4.94VSFGIHH139 pKa = 5.98NGEE142 pKa = 4.46DD143 pKa = 2.98ATIRR147 pKa = 11.84GDD149 pKa = 3.87FFGFTYY155 pKa = 10.56FGGVQVLEE163 pKa = 4.61GTAHH167 pKa = 6.58FSAGEE172 pKa = 3.85TDD174 pKa = 4.59FEE176 pKa = 4.94GFIYY180 pKa = 9.97WFNPAQTVRR189 pKa = 11.84GNARR193 pKa = 11.84LLGGDD198 pKa = 3.95DD199 pKa = 3.86TIIWTEE205 pKa = 3.93DD206 pKa = 2.98TRR208 pKa = 11.84SYY210 pKa = 10.24PSSYY214 pKa = 10.61VYY216 pKa = 10.57AYY218 pKa = 10.45ADD220 pKa = 3.28ASFVSGSAVVVGGDD234 pKa = 3.76DD235 pKa = 5.03LLDD238 pKa = 3.75ARR240 pKa = 11.84NAGDD244 pKa = 4.13GIGAFLVGDD253 pKa = 4.66AYY255 pKa = 10.28HH256 pKa = 5.93TTAATAMVIGGDD268 pKa = 4.37DD269 pKa = 3.34ILFGANASDD278 pKa = 3.72GAPTNRR284 pKa = 11.84LYY286 pKa = 11.53GDD288 pKa = 3.58VVVNFTDD295 pKa = 3.52TLIAGDD301 pKa = 3.56DD302 pKa = 4.04TIYY305 pKa = 11.19GRR307 pKa = 11.84AGNDD311 pKa = 3.26EE312 pKa = 4.47IYY314 pKa = 10.88GDD316 pKa = 3.89TGSTGTNYY324 pKa = 9.94IGGDD328 pKa = 3.45DD329 pKa = 5.21LIYY332 pKa = 10.79GGSDD336 pKa = 3.9GEE338 pKa = 4.11DD339 pKa = 2.99LMYY342 pKa = 10.77GGRR345 pKa = 11.84GNDD348 pKa = 2.63RR349 pKa = 11.84MYY351 pKa = 10.69GGKK354 pKa = 10.46GIVEE358 pKa = 3.94MRR360 pKa = 11.84GDD362 pKa = 3.47QDD364 pKa = 3.49NDD366 pKa = 3.03KK367 pKa = 10.84LYY369 pKa = 11.04AGDD372 pKa = 4.04GTTTAYY378 pKa = 10.36GGSGDD383 pKa = 5.14DD384 pKa = 4.38LIKK387 pKa = 10.51VTGDD391 pKa = 3.59GEE393 pKa = 4.62GSFYY397 pKa = 10.84GGSGIDD403 pKa = 3.27RR404 pKa = 11.84ISYY407 pKa = 9.77YY408 pKa = 10.46DD409 pKa = 3.56AEE411 pKa = 4.63GGITIDD417 pKa = 4.51LGANTISRR425 pKa = 11.84YY426 pKa = 8.71WADD429 pKa = 4.94DD430 pKa = 3.37DD431 pKa = 4.82TISNFEE437 pKa = 4.15NVSGSKK443 pKa = 10.23VGNNKK448 pKa = 9.17IFGTSGANDD457 pKa = 3.28IDD459 pKa = 4.27TYY461 pKa = 11.48DD462 pKa = 4.35GNDD465 pKa = 3.06KK466 pKa = 11.11VYY468 pKa = 11.06GGGGGDD474 pKa = 4.29KK475 pKa = 10.53ISLGKK480 pKa = 10.88GNDD483 pKa = 3.36YY484 pKa = 11.24VKK486 pKa = 11.14ADD488 pKa = 4.64FGANTYY494 pKa = 9.7EE495 pKa = 4.44GSSGLDD501 pKa = 3.03TISYY505 pKa = 9.69YY506 pKa = 11.05NSNDD510 pKa = 3.11GVIVDD515 pKa = 4.74LKK517 pKa = 10.96HH518 pKa = 6.39NEE520 pKa = 4.0AEE522 pKa = 4.17GGYY525 pKa = 10.79AEE527 pKa = 5.58GDD529 pKa = 3.7TISGFEE535 pKa = 4.19RR536 pKa = 11.84LTGSTEE542 pKa = 3.77DD543 pKa = 3.63DD544 pKa = 3.49KK545 pKa = 11.89LYY547 pKa = 11.16GSDD550 pKa = 4.12GGNILKK556 pKa = 10.52GGSGEE561 pKa = 3.99DD562 pKa = 3.8SLWGRR567 pKa = 11.84DD568 pKa = 4.05GNDD571 pKa = 3.07KK572 pKa = 11.09LYY574 pKa = 11.13GGSNSDD580 pKa = 3.46RR581 pKa = 11.84FDD583 pKa = 3.63GGEE586 pKa = 4.38GNDD589 pKa = 3.7KK590 pKa = 10.91LYY592 pKa = 11.14GGSGVDD598 pKa = 3.11TFHH601 pKa = 7.45FDD603 pKa = 4.17KK604 pKa = 11.3GDD606 pKa = 3.95DD607 pKa = 3.8FDD609 pKa = 4.83TVMDD613 pKa = 4.23FQNNADD619 pKa = 4.43RR620 pKa = 11.84IEE622 pKa = 4.45IDD624 pKa = 4.06GYY626 pKa = 11.52SSGFDD631 pKa = 2.99AFDD634 pKa = 3.57YY635 pKa = 8.49ATQVGTDD642 pKa = 3.2VVFNFGGGDD651 pKa = 3.58GLIVEE656 pKa = 4.81NVLIGQLGNDD666 pKa = 3.88LFVVV670 pKa = 4.15
Molecular weight: 70.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0S0D7|A0A1I0S0D7_9RHOB S-adenosylmethionine uptake transporter OS=Cognatiyoonia koreensis OX=364200 GN=SAMN04488515_3650 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.53ILNNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.47GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.53ILNNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.47GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1123027 |
31 |
4083 |
313.5 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.705 ± 0.057 | 0.911 ± 0.013 |
6.572 ± 0.049 | 5.505 ± 0.036 |
3.95 ± 0.029 | 8.579 ± 0.054 |
2.007 ± 0.024 | 5.631 ± 0.032 |
3.454 ± 0.037 | 9.55 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.838 ± 0.026 | 3.001 ± 0.029 |
4.874 ± 0.033 | 3.288 ± 0.024 |
5.952 ± 0.037 | 5.192 ± 0.028 |
5.95 ± 0.046 | 7.347 ± 0.033 |
1.376 ± 0.019 | 2.318 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
