
Horseradish latent virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Caulimovirus
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
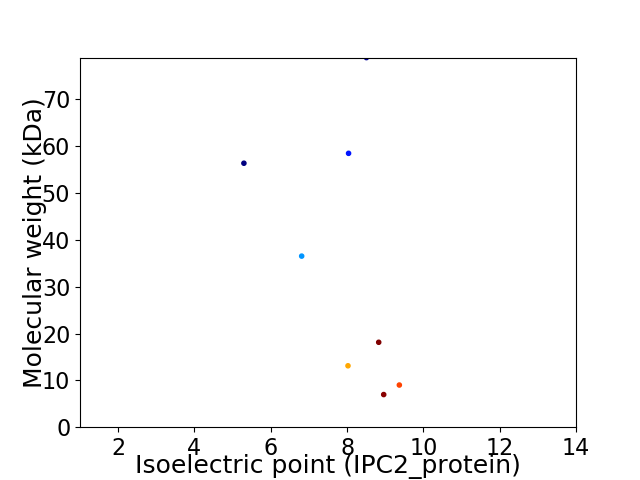
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q5J1S3|Q5J1S3_9VIRU Protein 3 OS=Horseradish latent virus OX=264076 PE=3 SV=1
MM1 pKa = 7.6PRR3 pKa = 11.84TVGQTIDD10 pKa = 5.02DD11 pKa = 4.01FWSNLGVNQLDD22 pKa = 3.57EE23 pKa = 5.06HH24 pKa = 6.57EE25 pKa = 4.72FDD27 pKa = 5.43LMINLMTDD35 pKa = 3.52DD36 pKa = 5.29DD37 pKa = 4.74VSSDD41 pKa = 3.35TSFDD45 pKa = 3.53SLTSLFSEE53 pKa = 4.72LLQVEE58 pKa = 4.18QEE60 pKa = 4.02ITSEE64 pKa = 4.09EE65 pKa = 4.3EE66 pKa = 3.72RR67 pKa = 11.84CQLLEE72 pKa = 4.96KK73 pKa = 10.21EE74 pKa = 4.54DD75 pKa = 4.37SSDD78 pKa = 3.25EE79 pKa = 4.48SIPNDD84 pKa = 3.34SDD86 pKa = 3.65EE87 pKa = 4.5EE88 pKa = 4.48SVPEE92 pKa = 3.85QVRR95 pKa = 11.84MEE97 pKa = 4.26KK98 pKa = 10.75GKK100 pKa = 11.12GPAKK104 pKa = 10.24PYY106 pKa = 10.65DD107 pKa = 3.69YY108 pKa = 10.89FDD110 pKa = 4.44NEE112 pKa = 4.37SSDD115 pKa = 3.75MEE117 pKa = 4.4YY118 pKa = 11.37GDD120 pKa = 3.98YY121 pKa = 11.13NPYY124 pKa = 10.02GRR126 pKa = 11.84SGQTNKK132 pKa = 9.93PARR135 pKa = 11.84TRR137 pKa = 11.84PKK139 pKa = 10.21LASEE143 pKa = 4.1TLGRR147 pKa = 11.84NPRR150 pKa = 11.84ILNLDD155 pKa = 3.64CTNSLRR161 pKa = 11.84DD162 pKa = 3.36RR163 pKa = 11.84KK164 pKa = 10.38RR165 pKa = 11.84LVEE168 pKa = 3.85EE169 pKa = 4.12WGAEE173 pKa = 3.52ISLTIQTNLDD183 pKa = 3.83EE184 pKa = 5.12YY185 pKa = 11.1SDD187 pKa = 4.16PDD189 pKa = 3.6MVLLLMEE196 pKa = 5.22HH197 pKa = 6.42LTAGSVKK204 pKa = 10.66SFIKK208 pKa = 6.67TTKK211 pKa = 9.21WSEE214 pKa = 3.94LSGGIYY220 pKa = 10.23DD221 pKa = 4.44LVMEE225 pKa = 6.0GIHH228 pKa = 7.11VMFLGEE234 pKa = 4.06QPNEE238 pKa = 4.04AFDD241 pKa = 3.79KK242 pKa = 11.12AKK244 pKa = 10.29EE245 pKa = 3.63QAAAKK250 pKa = 10.23DD251 pKa = 3.91KK252 pKa = 10.58LLKK255 pKa = 9.68MQLCDD260 pKa = 3.12ICSLDD265 pKa = 3.39TFTCAFEE272 pKa = 3.96KK273 pKa = 10.77ALYY276 pKa = 10.25KK277 pKa = 10.3LDD279 pKa = 4.4SGDD282 pKa = 3.73FPPIIEE288 pKa = 4.39QYY290 pKa = 9.43LAKK293 pKa = 10.17IPEE296 pKa = 4.34VGAKK300 pKa = 9.21AQQRR304 pKa = 11.84YY305 pKa = 9.03KK306 pKa = 11.06EE307 pKa = 4.13EE308 pKa = 3.72ATGAMKK314 pKa = 10.83YY315 pKa = 10.76SLGFANKK322 pKa = 9.23IVKK325 pKa = 10.23EE326 pKa = 3.93EE327 pKa = 3.96LKK329 pKa = 10.6KK330 pKa = 10.55VCEE333 pKa = 4.03LTRR336 pKa = 11.84LQKK339 pKa = 10.53KK340 pKa = 9.31LKK342 pKa = 10.06KK343 pKa = 9.95FRR345 pKa = 11.84KK346 pKa = 6.93TCCKK350 pKa = 9.74QVEE353 pKa = 4.15EE354 pKa = 4.15RR355 pKa = 11.84LEE357 pKa = 4.1YY358 pKa = 10.59GCRR361 pKa = 11.84PIYY364 pKa = 9.89KK365 pKa = 9.64KK366 pKa = 10.17KK367 pKa = 10.16SKK369 pKa = 9.87RR370 pKa = 11.84RR371 pKa = 11.84KK372 pKa = 8.48PKK374 pKa = 8.02VRR376 pKa = 11.84KK377 pKa = 7.87YY378 pKa = 10.95KK379 pKa = 10.16KK380 pKa = 8.97YY381 pKa = 10.74KK382 pKa = 8.28PFKK385 pKa = 9.51RR386 pKa = 11.84RR387 pKa = 11.84KK388 pKa = 8.41KK389 pKa = 10.35FKK391 pKa = 7.66TGKK394 pKa = 8.91YY395 pKa = 9.12FRR397 pKa = 11.84PKK399 pKa = 9.62TEE401 pKa = 4.31KK402 pKa = 10.41KK403 pKa = 10.04VCPKK407 pKa = 10.94GKK409 pKa = 10.36GKK411 pKa = 9.66NCKK414 pKa = 9.48CWICNVEE421 pKa = 3.58GHH423 pKa = 6.09YY424 pKa = 11.32ANEE427 pKa = 4.32CPNRR431 pKa = 11.84QTSQKK436 pKa = 10.95AHH438 pKa = 6.98ILQEE442 pKa = 4.56AIQLGLQPVEE452 pKa = 4.2DD453 pKa = 4.23VYY455 pKa = 11.42EE456 pKa = 4.67GEE458 pKa = 4.1QEE460 pKa = 4.3VFVIEE465 pKa = 4.26YY466 pKa = 10.22IEE468 pKa = 4.09VSEE471 pKa = 5.27DD472 pKa = 3.36EE473 pKa = 4.57TSTSEE478 pKa = 4.98DD479 pKa = 3.73DD480 pKa = 3.79GTSTSEE486 pKa = 5.44DD487 pKa = 3.49SDD489 pKa = 3.85SDD491 pKa = 3.58
MM1 pKa = 7.6PRR3 pKa = 11.84TVGQTIDD10 pKa = 5.02DD11 pKa = 4.01FWSNLGVNQLDD22 pKa = 3.57EE23 pKa = 5.06HH24 pKa = 6.57EE25 pKa = 4.72FDD27 pKa = 5.43LMINLMTDD35 pKa = 3.52DD36 pKa = 5.29DD37 pKa = 4.74VSSDD41 pKa = 3.35TSFDD45 pKa = 3.53SLTSLFSEE53 pKa = 4.72LLQVEE58 pKa = 4.18QEE60 pKa = 4.02ITSEE64 pKa = 4.09EE65 pKa = 4.3EE66 pKa = 3.72RR67 pKa = 11.84CQLLEE72 pKa = 4.96KK73 pKa = 10.21EE74 pKa = 4.54DD75 pKa = 4.37SSDD78 pKa = 3.25EE79 pKa = 4.48SIPNDD84 pKa = 3.34SDD86 pKa = 3.65EE87 pKa = 4.5EE88 pKa = 4.48SVPEE92 pKa = 3.85QVRR95 pKa = 11.84MEE97 pKa = 4.26KK98 pKa = 10.75GKK100 pKa = 11.12GPAKK104 pKa = 10.24PYY106 pKa = 10.65DD107 pKa = 3.69YY108 pKa = 10.89FDD110 pKa = 4.44NEE112 pKa = 4.37SSDD115 pKa = 3.75MEE117 pKa = 4.4YY118 pKa = 11.37GDD120 pKa = 3.98YY121 pKa = 11.13NPYY124 pKa = 10.02GRR126 pKa = 11.84SGQTNKK132 pKa = 9.93PARR135 pKa = 11.84TRR137 pKa = 11.84PKK139 pKa = 10.21LASEE143 pKa = 4.1TLGRR147 pKa = 11.84NPRR150 pKa = 11.84ILNLDD155 pKa = 3.64CTNSLRR161 pKa = 11.84DD162 pKa = 3.36RR163 pKa = 11.84KK164 pKa = 10.38RR165 pKa = 11.84LVEE168 pKa = 3.85EE169 pKa = 4.12WGAEE173 pKa = 3.52ISLTIQTNLDD183 pKa = 3.83EE184 pKa = 5.12YY185 pKa = 11.1SDD187 pKa = 4.16PDD189 pKa = 3.6MVLLLMEE196 pKa = 5.22HH197 pKa = 6.42LTAGSVKK204 pKa = 10.66SFIKK208 pKa = 6.67TTKK211 pKa = 9.21WSEE214 pKa = 3.94LSGGIYY220 pKa = 10.23DD221 pKa = 4.44LVMEE225 pKa = 6.0GIHH228 pKa = 7.11VMFLGEE234 pKa = 4.06QPNEE238 pKa = 4.04AFDD241 pKa = 3.79KK242 pKa = 11.12AKK244 pKa = 10.29EE245 pKa = 3.63QAAAKK250 pKa = 10.23DD251 pKa = 3.91KK252 pKa = 10.58LLKK255 pKa = 9.68MQLCDD260 pKa = 3.12ICSLDD265 pKa = 3.39TFTCAFEE272 pKa = 3.96KK273 pKa = 10.77ALYY276 pKa = 10.25KK277 pKa = 10.3LDD279 pKa = 4.4SGDD282 pKa = 3.73FPPIIEE288 pKa = 4.39QYY290 pKa = 9.43LAKK293 pKa = 10.17IPEE296 pKa = 4.34VGAKK300 pKa = 9.21AQQRR304 pKa = 11.84YY305 pKa = 9.03KK306 pKa = 11.06EE307 pKa = 4.13EE308 pKa = 3.72ATGAMKK314 pKa = 10.83YY315 pKa = 10.76SLGFANKK322 pKa = 9.23IVKK325 pKa = 10.23EE326 pKa = 3.93EE327 pKa = 3.96LKK329 pKa = 10.6KK330 pKa = 10.55VCEE333 pKa = 4.03LTRR336 pKa = 11.84LQKK339 pKa = 10.53KK340 pKa = 9.31LKK342 pKa = 10.06KK343 pKa = 9.95FRR345 pKa = 11.84KK346 pKa = 6.93TCCKK350 pKa = 9.74QVEE353 pKa = 4.15EE354 pKa = 4.15RR355 pKa = 11.84LEE357 pKa = 4.1YY358 pKa = 10.59GCRR361 pKa = 11.84PIYY364 pKa = 9.89KK365 pKa = 9.64KK366 pKa = 10.17KK367 pKa = 10.16SKK369 pKa = 9.87RR370 pKa = 11.84RR371 pKa = 11.84KK372 pKa = 8.48PKK374 pKa = 8.02VRR376 pKa = 11.84KK377 pKa = 7.87YY378 pKa = 10.95KK379 pKa = 10.16KK380 pKa = 8.97YY381 pKa = 10.74KK382 pKa = 8.28PFKK385 pKa = 9.51RR386 pKa = 11.84RR387 pKa = 11.84KK388 pKa = 8.41KK389 pKa = 10.35FKK391 pKa = 7.66TGKK394 pKa = 8.91YY395 pKa = 9.12FRR397 pKa = 11.84PKK399 pKa = 9.62TEE401 pKa = 4.31KK402 pKa = 10.41KK403 pKa = 10.04VCPKK407 pKa = 10.94GKK409 pKa = 10.36GKK411 pKa = 9.66NCKK414 pKa = 9.48CWICNVEE421 pKa = 3.58GHH423 pKa = 6.09YY424 pKa = 11.32ANEE427 pKa = 4.32CPNRR431 pKa = 11.84QTSQKK436 pKa = 10.95AHH438 pKa = 6.98ILQEE442 pKa = 4.56AIQLGLQPVEE452 pKa = 4.2DD453 pKa = 4.23VYY455 pKa = 11.42EE456 pKa = 4.67GEE458 pKa = 4.1QEE460 pKa = 4.3VFVIEE465 pKa = 4.26YY466 pKa = 10.22IEE468 pKa = 4.09VSEE471 pKa = 5.27DD472 pKa = 3.36EE473 pKa = 4.57TSTSEE478 pKa = 4.98DD479 pKa = 3.73DD480 pKa = 3.79GTSTSEE486 pKa = 5.44DD487 pKa = 3.49SDD489 pKa = 3.85SDD491 pKa = 3.58
Molecular weight: 56.37 kDa
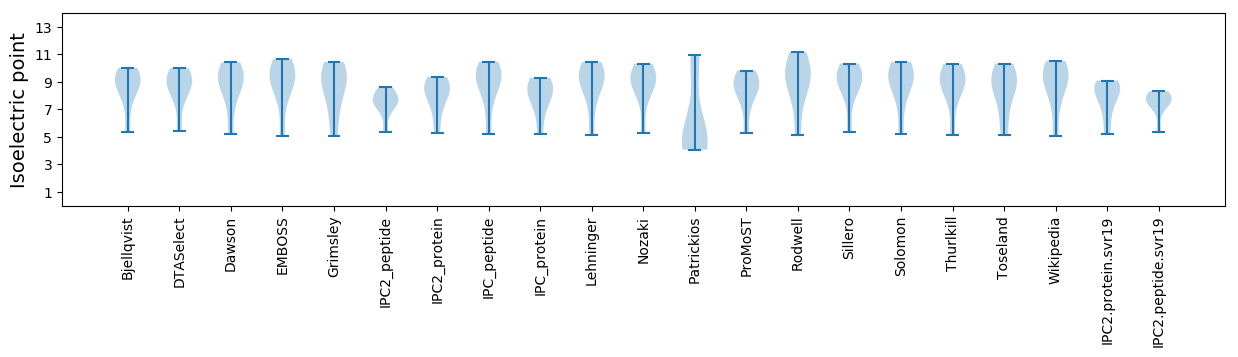
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0DKX7|K0DKX7_9VIRU Uncharacterized protein OS=Horseradish latent virus OX=264076 PE=4 SV=1
MM1 pKa = 7.84KK2 pKa = 10.41DD3 pKa = 3.23NEE5 pKa = 4.52KK6 pKa = 10.02TGSSDD11 pKa = 3.37GHH13 pKa = 6.82CSGQIYY19 pKa = 10.81LNLLILSLRR28 pKa = 11.84RR29 pKa = 11.84LTLISGSKK37 pKa = 10.4GLLINPYY44 pKa = 7.57QFKK47 pKa = 10.6ILLIKK52 pKa = 10.23KK53 pKa = 8.28LNKK56 pKa = 8.75WFKK59 pKa = 9.05CHH61 pKa = 6.8KK62 pKa = 8.93STFRR66 pKa = 11.84STNLVVQKK74 pKa = 10.6LVSEE78 pKa = 4.52PP79 pKa = 3.46
MM1 pKa = 7.84KK2 pKa = 10.41DD3 pKa = 3.23NEE5 pKa = 4.52KK6 pKa = 10.02TGSSDD11 pKa = 3.37GHH13 pKa = 6.82CSGQIYY19 pKa = 10.81LNLLILSLRR28 pKa = 11.84RR29 pKa = 11.84LTLISGSKK37 pKa = 10.4GLLINPYY44 pKa = 7.57QFKK47 pKa = 10.6ILLIKK52 pKa = 10.23KK53 pKa = 8.28LNKK56 pKa = 8.75WFKK59 pKa = 9.05CHH61 pKa = 6.8KK62 pKa = 8.93STFRR66 pKa = 11.84STNLVVQKK74 pKa = 10.6LVSEE78 pKa = 4.52PP79 pKa = 3.46
Molecular weight: 9.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2435 |
60 |
679 |
304.4 |
34.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.6 ± 0.489 | 1.889 ± 0.255 |
5.298 ± 0.459 | 7.885 ± 0.719 |
3.901 ± 0.493 | 4.969 ± 0.19 |
1.725 ± 0.365 | 7.351 ± 0.72 |
10.554 ± 0.551 | 8.665 ± 0.647 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.177 ± 0.194 | 5.421 ± 0.623 |
4.6 ± 0.519 | 4.23 ± 0.493 |
4.312 ± 0.409 | 7.598 ± 0.717 |
5.955 ± 0.563 | 4.887 ± 0.202 |
0.821 ± 0.173 | 3.162 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
