
Botrytis cinerea mitovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
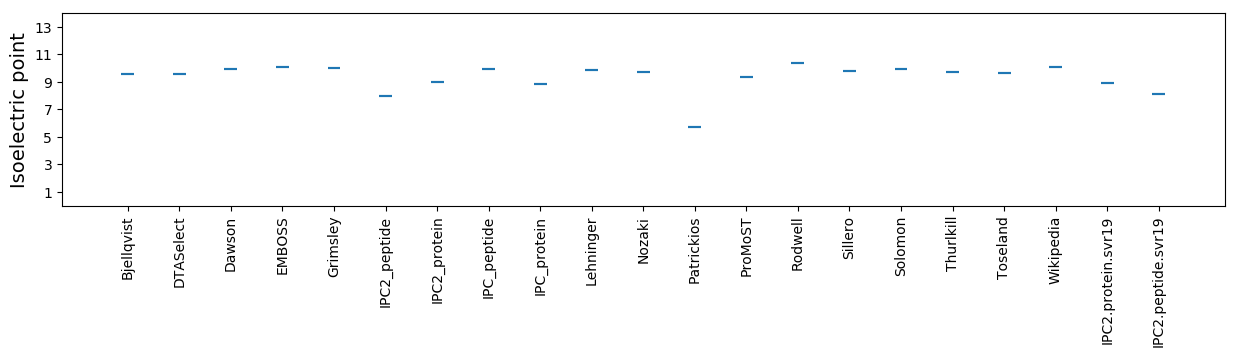
Average proteome isoelectric point is 8.92
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S4GA49|A0A0S4GA49_9VIRU RNA dependent RNA polymerase OS=Botrytis cinerea mitovirus 2 OX=1629665 GN=RdRp PE=4 SV=1
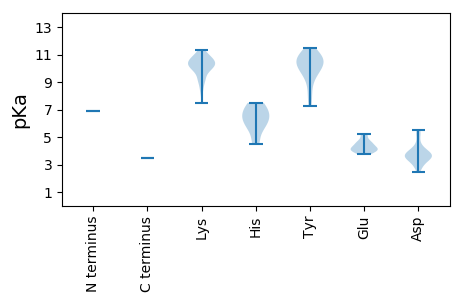
MM1 pKa = 6.92KK2 pKa = 10.18TKK4 pKa = 10.51FLVTKK9 pKa = 10.34RR10 pKa = 11.84LLEE13 pKa = 4.06SLFDD17 pKa = 4.36DD18 pKa = 3.52RR19 pKa = 11.84TIGKK23 pKa = 9.75KK24 pKa = 9.99YY25 pKa = 9.17IKK27 pKa = 10.22SVEE30 pKa = 3.95TMRR33 pKa = 11.84IKK35 pKa = 10.68SGLPFTIKK43 pKa = 10.49YY44 pKa = 7.28MKK46 pKa = 10.17AVKK49 pKa = 10.12LHH51 pKa = 4.48ITRR54 pKa = 11.84YY55 pKa = 9.89ISGQPLRR62 pKa = 11.84TNSSLVSLTNGFPTKK77 pKa = 10.49FLYY80 pKa = 10.5LKK82 pKa = 10.23EE83 pKa = 5.23LIDD86 pKa = 4.0SGDD89 pKa = 4.0PIKK92 pKa = 10.93LRR94 pKa = 11.84LVLTLTGYY102 pKa = 8.31TRR104 pKa = 11.84SIIPTKK110 pKa = 10.75DD111 pKa = 2.89EE112 pKa = 4.11EE113 pKa = 4.57KK114 pKa = 10.64LVKK117 pKa = 9.05PTFNSISDD125 pKa = 3.82PYY127 pKa = 10.16KK128 pKa = 10.22GKK130 pKa = 10.63EE131 pKa = 3.81NYY133 pKa = 7.81TIPTEE138 pKa = 4.53FINGFVEE145 pKa = 4.61KK146 pKa = 10.67YY147 pKa = 10.17KK148 pKa = 11.15LNFMPKK154 pKa = 8.37WDD156 pKa = 4.33NSLQYY161 pKa = 10.31ISNKK165 pKa = 9.71SSPFGKK171 pKa = 8.51STLTGPFALFHH182 pKa = 5.94MGHH185 pKa = 6.97WNLNMLDD192 pKa = 3.99HH193 pKa = 6.77FKK195 pKa = 11.33SLIGDD200 pKa = 4.12GSYY203 pKa = 10.93KK204 pKa = 10.54IMIGEE209 pKa = 4.02LLEE212 pKa = 4.99NVFKK216 pKa = 10.7DD217 pKa = 3.43HH218 pKa = 7.37RR219 pKa = 11.84CFHH222 pKa = 7.0IGNSLNGIGKK232 pKa = 9.06IAIVKK237 pKa = 10.29DD238 pKa = 3.58PEE240 pKa = 4.14LKK242 pKa = 10.01QRR244 pKa = 11.84AIAMVDD250 pKa = 3.61YY251 pKa = 10.72YY252 pKa = 11.06SQLVLRR258 pKa = 11.84PIHH261 pKa = 7.41DD262 pKa = 3.87GLLNKK267 pKa = 9.71LRR269 pKa = 11.84NLPQDD274 pKa = 3.11RR275 pKa = 11.84TFTQNPFNNWGKK287 pKa = 8.89TMGNSFWSLDD297 pKa = 3.57LSSATDD303 pKa = 3.75RR304 pKa = 11.84FPISLQEE311 pKa = 4.09KK312 pKa = 9.2VLASILKK319 pKa = 10.35DD320 pKa = 3.34EE321 pKa = 5.22AKK323 pKa = 10.82AEE325 pKa = 4.17TWRR328 pKa = 11.84KK329 pKa = 9.25ILIEE333 pKa = 4.31RR334 pKa = 11.84DD335 pKa = 3.38YY336 pKa = 11.49KK337 pKa = 11.09LPNGNLTRR345 pKa = 11.84YY346 pKa = 9.85SVGQPMGAYY355 pKa = 10.07SSWTAFTISHH365 pKa = 6.51HH366 pKa = 6.03LVVHH370 pKa = 6.49FAAHH374 pKa = 6.48LCGIEE379 pKa = 4.64NFDD382 pKa = 4.69KK383 pKa = 11.23YY384 pKa = 11.07ILLGDD389 pKa = 5.1DD390 pKa = 3.46IVINNDD396 pKa = 2.48KK397 pKa = 10.69VARR400 pKa = 11.84KK401 pKa = 9.27YY402 pKa = 10.71IKK404 pKa = 10.28IMTKK408 pKa = 10.23LGVDD412 pKa = 3.21ISINKK417 pKa = 7.45THH419 pKa = 5.9VSKK422 pKa = 9.84NTYY425 pKa = 9.41EE426 pKa = 4.06FAKK429 pKa = 10.2RR430 pKa = 11.84WVKK433 pKa = 10.74HH434 pKa = 5.65NIEE437 pKa = 3.86ITGIPLRR444 pKa = 11.84GILNNLNSLPTAIKK458 pKa = 9.31QLVYY462 pKa = 10.79YY463 pKa = 9.58FYY465 pKa = 11.34SCNTLWRR472 pKa = 11.84GNSVEE477 pKa = 4.75LIFSIFKK484 pKa = 10.15DD485 pKa = 2.82IKK487 pKa = 10.11LRR489 pKa = 11.84NRR491 pKa = 11.84FITDD495 pKa = 2.79SKK497 pKa = 11.3LRR499 pKa = 11.84ILSEE503 pKa = 3.74DD504 pKa = 3.64TIFVIRR510 pKa = 11.84QTLKK514 pKa = 10.12LNTYY518 pKa = 8.86EE519 pKa = 4.53EE520 pKa = 3.86IRR522 pKa = 11.84NYY524 pKa = 10.08FNKK527 pKa = 10.36KK528 pKa = 8.2LTIEE532 pKa = 4.08NVLIPNEE539 pKa = 4.01DD540 pKa = 3.62QIHH543 pKa = 5.4GFIRR547 pKa = 11.84VILCLGLNKK556 pKa = 9.82VAEE559 pKa = 4.25KK560 pKa = 10.72SGNDD564 pKa = 3.02LSKK567 pKa = 10.75YY568 pKa = 9.72YY569 pKa = 10.82KK570 pKa = 10.54SFEE573 pKa = 4.18KK574 pKa = 10.68NFRR577 pKa = 11.84KK578 pKa = 10.26GDD580 pKa = 3.59FDD582 pKa = 5.52LKK584 pKa = 10.38MLVRR588 pKa = 11.84HH589 pKa = 6.45PIVLGIYY596 pKa = 10.38NKK598 pKa = 9.63ISQMKK603 pKa = 8.8RR604 pKa = 11.84TLQKK608 pKa = 9.83VRR610 pKa = 11.84QMDD613 pKa = 3.9EE614 pKa = 4.36LDD616 pKa = 5.23LIDD619 pKa = 5.5AMTHH623 pKa = 4.67MRR625 pKa = 11.84IDD627 pKa = 3.78EE628 pKa = 4.44PDD630 pKa = 3.55KK631 pKa = 11.07LVQSLRR637 pKa = 11.84NTSRR641 pKa = 11.84SISHH645 pKa = 7.49IDD647 pKa = 3.52QLWKK651 pKa = 10.7KK652 pKa = 9.32GFKK655 pKa = 9.98RR656 pKa = 11.84LEE658 pKa = 4.35IINEE662 pKa = 3.83DD663 pKa = 3.4NYY665 pKa = 10.88VNFNEE670 pKa = 5.14LDD672 pKa = 3.78LPNKK676 pKa = 10.31NNLKK680 pKa = 9.79PWEE683 pKa = 4.22SYY685 pKa = 10.64YY686 pKa = 10.31ISNLSEE692 pKa = 4.74LSDD695 pKa = 3.97KK696 pKa = 11.0LDD698 pKa = 3.58SLKK701 pKa = 10.14TPVTQNMWFF710 pKa = 3.48
MM1 pKa = 6.92KK2 pKa = 10.18TKK4 pKa = 10.51FLVTKK9 pKa = 10.34RR10 pKa = 11.84LLEE13 pKa = 4.06SLFDD17 pKa = 4.36DD18 pKa = 3.52RR19 pKa = 11.84TIGKK23 pKa = 9.75KK24 pKa = 9.99YY25 pKa = 9.17IKK27 pKa = 10.22SVEE30 pKa = 3.95TMRR33 pKa = 11.84IKK35 pKa = 10.68SGLPFTIKK43 pKa = 10.49YY44 pKa = 7.28MKK46 pKa = 10.17AVKK49 pKa = 10.12LHH51 pKa = 4.48ITRR54 pKa = 11.84YY55 pKa = 9.89ISGQPLRR62 pKa = 11.84TNSSLVSLTNGFPTKK77 pKa = 10.49FLYY80 pKa = 10.5LKK82 pKa = 10.23EE83 pKa = 5.23LIDD86 pKa = 4.0SGDD89 pKa = 4.0PIKK92 pKa = 10.93LRR94 pKa = 11.84LVLTLTGYY102 pKa = 8.31TRR104 pKa = 11.84SIIPTKK110 pKa = 10.75DD111 pKa = 2.89EE112 pKa = 4.11EE113 pKa = 4.57KK114 pKa = 10.64LVKK117 pKa = 9.05PTFNSISDD125 pKa = 3.82PYY127 pKa = 10.16KK128 pKa = 10.22GKK130 pKa = 10.63EE131 pKa = 3.81NYY133 pKa = 7.81TIPTEE138 pKa = 4.53FINGFVEE145 pKa = 4.61KK146 pKa = 10.67YY147 pKa = 10.17KK148 pKa = 11.15LNFMPKK154 pKa = 8.37WDD156 pKa = 4.33NSLQYY161 pKa = 10.31ISNKK165 pKa = 9.71SSPFGKK171 pKa = 8.51STLTGPFALFHH182 pKa = 5.94MGHH185 pKa = 6.97WNLNMLDD192 pKa = 3.99HH193 pKa = 6.77FKK195 pKa = 11.33SLIGDD200 pKa = 4.12GSYY203 pKa = 10.93KK204 pKa = 10.54IMIGEE209 pKa = 4.02LLEE212 pKa = 4.99NVFKK216 pKa = 10.7DD217 pKa = 3.43HH218 pKa = 7.37RR219 pKa = 11.84CFHH222 pKa = 7.0IGNSLNGIGKK232 pKa = 9.06IAIVKK237 pKa = 10.29DD238 pKa = 3.58PEE240 pKa = 4.14LKK242 pKa = 10.01QRR244 pKa = 11.84AIAMVDD250 pKa = 3.61YY251 pKa = 10.72YY252 pKa = 11.06SQLVLRR258 pKa = 11.84PIHH261 pKa = 7.41DD262 pKa = 3.87GLLNKK267 pKa = 9.71LRR269 pKa = 11.84NLPQDD274 pKa = 3.11RR275 pKa = 11.84TFTQNPFNNWGKK287 pKa = 8.89TMGNSFWSLDD297 pKa = 3.57LSSATDD303 pKa = 3.75RR304 pKa = 11.84FPISLQEE311 pKa = 4.09KK312 pKa = 9.2VLASILKK319 pKa = 10.35DD320 pKa = 3.34EE321 pKa = 5.22AKK323 pKa = 10.82AEE325 pKa = 4.17TWRR328 pKa = 11.84KK329 pKa = 9.25ILIEE333 pKa = 4.31RR334 pKa = 11.84DD335 pKa = 3.38YY336 pKa = 11.49KK337 pKa = 11.09LPNGNLTRR345 pKa = 11.84YY346 pKa = 9.85SVGQPMGAYY355 pKa = 10.07SSWTAFTISHH365 pKa = 6.51HH366 pKa = 6.03LVVHH370 pKa = 6.49FAAHH374 pKa = 6.48LCGIEE379 pKa = 4.64NFDD382 pKa = 4.69KK383 pKa = 11.23YY384 pKa = 11.07ILLGDD389 pKa = 5.1DD390 pKa = 3.46IVINNDD396 pKa = 2.48KK397 pKa = 10.69VARR400 pKa = 11.84KK401 pKa = 9.27YY402 pKa = 10.71IKK404 pKa = 10.28IMTKK408 pKa = 10.23LGVDD412 pKa = 3.21ISINKK417 pKa = 7.45THH419 pKa = 5.9VSKK422 pKa = 9.84NTYY425 pKa = 9.41EE426 pKa = 4.06FAKK429 pKa = 10.2RR430 pKa = 11.84WVKK433 pKa = 10.74HH434 pKa = 5.65NIEE437 pKa = 3.86ITGIPLRR444 pKa = 11.84GILNNLNSLPTAIKK458 pKa = 9.31QLVYY462 pKa = 10.79YY463 pKa = 9.58FYY465 pKa = 11.34SCNTLWRR472 pKa = 11.84GNSVEE477 pKa = 4.75LIFSIFKK484 pKa = 10.15DD485 pKa = 2.82IKK487 pKa = 10.11LRR489 pKa = 11.84NRR491 pKa = 11.84FITDD495 pKa = 2.79SKK497 pKa = 11.3LRR499 pKa = 11.84ILSEE503 pKa = 3.74DD504 pKa = 3.64TIFVIRR510 pKa = 11.84QTLKK514 pKa = 10.12LNTYY518 pKa = 8.86EE519 pKa = 4.53EE520 pKa = 3.86IRR522 pKa = 11.84NYY524 pKa = 10.08FNKK527 pKa = 10.36KK528 pKa = 8.2LTIEE532 pKa = 4.08NVLIPNEE539 pKa = 4.01DD540 pKa = 3.62QIHH543 pKa = 5.4GFIRR547 pKa = 11.84VILCLGLNKK556 pKa = 9.82VAEE559 pKa = 4.25KK560 pKa = 10.72SGNDD564 pKa = 3.02LSKK567 pKa = 10.75YY568 pKa = 9.72YY569 pKa = 10.82KK570 pKa = 10.54SFEE573 pKa = 4.18KK574 pKa = 10.68NFRR577 pKa = 11.84KK578 pKa = 10.26GDD580 pKa = 3.59FDD582 pKa = 5.52LKK584 pKa = 10.38MLVRR588 pKa = 11.84HH589 pKa = 6.45PIVLGIYY596 pKa = 10.38NKK598 pKa = 9.63ISQMKK603 pKa = 8.8RR604 pKa = 11.84TLQKK608 pKa = 9.83VRR610 pKa = 11.84QMDD613 pKa = 3.9EE614 pKa = 4.36LDD616 pKa = 5.23LIDD619 pKa = 5.5AMTHH623 pKa = 4.67MRR625 pKa = 11.84IDD627 pKa = 3.78EE628 pKa = 4.44PDD630 pKa = 3.55KK631 pKa = 11.07LVQSLRR637 pKa = 11.84NTSRR641 pKa = 11.84SISHH645 pKa = 7.49IDD647 pKa = 3.52QLWKK651 pKa = 10.7KK652 pKa = 9.32GFKK655 pKa = 9.98RR656 pKa = 11.84LEE658 pKa = 4.35IINEE662 pKa = 3.83DD663 pKa = 3.4NYY665 pKa = 10.88VNFNEE670 pKa = 5.14LDD672 pKa = 3.78LPNKK676 pKa = 10.31NNLKK680 pKa = 9.79PWEE683 pKa = 4.22SYY685 pKa = 10.64YY686 pKa = 10.31ISNLSEE692 pKa = 4.74LSDD695 pKa = 3.97KK696 pKa = 11.0LDD698 pKa = 3.58SLKK701 pKa = 10.14TPVTQNMWFF710 pKa = 3.48
Molecular weight: 82.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S4GA49|A0A0S4GA49_9VIRU RNA dependent RNA polymerase OS=Botrytis cinerea mitovirus 2 OX=1629665 GN=RdRp PE=4 SV=1
MM1 pKa = 6.92KK2 pKa = 10.18TKK4 pKa = 10.51FLVTKK9 pKa = 10.34RR10 pKa = 11.84LLEE13 pKa = 4.06SLFDD17 pKa = 4.36DD18 pKa = 3.52RR19 pKa = 11.84TIGKK23 pKa = 9.75KK24 pKa = 9.99YY25 pKa = 9.17IKK27 pKa = 10.22SVEE30 pKa = 3.95TMRR33 pKa = 11.84IKK35 pKa = 10.68SGLPFTIKK43 pKa = 10.49YY44 pKa = 7.28MKK46 pKa = 10.17AVKK49 pKa = 10.12LHH51 pKa = 4.48ITRR54 pKa = 11.84YY55 pKa = 9.89ISGQPLRR62 pKa = 11.84TNSSLVSLTNGFPTKK77 pKa = 10.49FLYY80 pKa = 10.5LKK82 pKa = 10.23EE83 pKa = 5.23LIDD86 pKa = 4.0SGDD89 pKa = 4.0PIKK92 pKa = 10.93LRR94 pKa = 11.84LVLTLTGYY102 pKa = 8.31TRR104 pKa = 11.84SIIPTKK110 pKa = 10.75DD111 pKa = 2.89EE112 pKa = 4.11EE113 pKa = 4.57KK114 pKa = 10.64LVKK117 pKa = 9.05PTFNSISDD125 pKa = 3.82PYY127 pKa = 10.16KK128 pKa = 10.22GKK130 pKa = 10.63EE131 pKa = 3.81NYY133 pKa = 7.81TIPTEE138 pKa = 4.53FINGFVEE145 pKa = 4.61KK146 pKa = 10.67YY147 pKa = 10.17KK148 pKa = 11.15LNFMPKK154 pKa = 8.37WDD156 pKa = 4.33NSLQYY161 pKa = 10.31ISNKK165 pKa = 9.71SSPFGKK171 pKa = 8.51STLTGPFALFHH182 pKa = 5.94MGHH185 pKa = 6.97WNLNMLDD192 pKa = 3.99HH193 pKa = 6.77FKK195 pKa = 11.33SLIGDD200 pKa = 4.12GSYY203 pKa = 10.93KK204 pKa = 10.54IMIGEE209 pKa = 4.02LLEE212 pKa = 4.99NVFKK216 pKa = 10.7DD217 pKa = 3.43HH218 pKa = 7.37RR219 pKa = 11.84CFHH222 pKa = 7.0IGNSLNGIGKK232 pKa = 9.06IAIVKK237 pKa = 10.29DD238 pKa = 3.58PEE240 pKa = 4.14LKK242 pKa = 10.01QRR244 pKa = 11.84AIAMVDD250 pKa = 3.61YY251 pKa = 10.72YY252 pKa = 11.06SQLVLRR258 pKa = 11.84PIHH261 pKa = 7.41DD262 pKa = 3.87GLLNKK267 pKa = 9.71LRR269 pKa = 11.84NLPQDD274 pKa = 3.11RR275 pKa = 11.84TFTQNPFNNWGKK287 pKa = 8.89TMGNSFWSLDD297 pKa = 3.57LSSATDD303 pKa = 3.75RR304 pKa = 11.84FPISLQEE311 pKa = 4.09KK312 pKa = 9.2VLASILKK319 pKa = 10.35DD320 pKa = 3.34EE321 pKa = 5.22AKK323 pKa = 10.82AEE325 pKa = 4.17TWRR328 pKa = 11.84KK329 pKa = 9.25ILIEE333 pKa = 4.31RR334 pKa = 11.84DD335 pKa = 3.38YY336 pKa = 11.49KK337 pKa = 11.09LPNGNLTRR345 pKa = 11.84YY346 pKa = 9.85SVGQPMGAYY355 pKa = 10.07SSWTAFTISHH365 pKa = 6.51HH366 pKa = 6.03LVVHH370 pKa = 6.49FAAHH374 pKa = 6.48LCGIEE379 pKa = 4.64NFDD382 pKa = 4.69KK383 pKa = 11.23YY384 pKa = 11.07ILLGDD389 pKa = 5.1DD390 pKa = 3.46IVINNDD396 pKa = 2.48KK397 pKa = 10.69VARR400 pKa = 11.84KK401 pKa = 9.27YY402 pKa = 10.71IKK404 pKa = 10.28IMTKK408 pKa = 10.23LGVDD412 pKa = 3.21ISINKK417 pKa = 7.45THH419 pKa = 5.9VSKK422 pKa = 9.84NTYY425 pKa = 9.41EE426 pKa = 4.06FAKK429 pKa = 10.2RR430 pKa = 11.84WVKK433 pKa = 10.74HH434 pKa = 5.65NIEE437 pKa = 3.86ITGIPLRR444 pKa = 11.84GILNNLNSLPTAIKK458 pKa = 9.31QLVYY462 pKa = 10.79YY463 pKa = 9.58FYY465 pKa = 11.34SCNTLWRR472 pKa = 11.84GNSVEE477 pKa = 4.75LIFSIFKK484 pKa = 10.15DD485 pKa = 2.82IKK487 pKa = 10.11LRR489 pKa = 11.84NRR491 pKa = 11.84FITDD495 pKa = 2.79SKK497 pKa = 11.3LRR499 pKa = 11.84ILSEE503 pKa = 3.74DD504 pKa = 3.64TIFVIRR510 pKa = 11.84QTLKK514 pKa = 10.12LNTYY518 pKa = 8.86EE519 pKa = 4.53EE520 pKa = 3.86IRR522 pKa = 11.84NYY524 pKa = 10.08FNKK527 pKa = 10.36KK528 pKa = 8.2LTIEE532 pKa = 4.08NVLIPNEE539 pKa = 4.01DD540 pKa = 3.62QIHH543 pKa = 5.4GFIRR547 pKa = 11.84VILCLGLNKK556 pKa = 9.82VAEE559 pKa = 4.25KK560 pKa = 10.72SGNDD564 pKa = 3.02LSKK567 pKa = 10.75YY568 pKa = 9.72YY569 pKa = 10.82KK570 pKa = 10.54SFEE573 pKa = 4.18KK574 pKa = 10.68NFRR577 pKa = 11.84KK578 pKa = 10.26GDD580 pKa = 3.59FDD582 pKa = 5.52LKK584 pKa = 10.38MLVRR588 pKa = 11.84HH589 pKa = 6.45PIVLGIYY596 pKa = 10.38NKK598 pKa = 9.63ISQMKK603 pKa = 8.8RR604 pKa = 11.84TLQKK608 pKa = 9.83VRR610 pKa = 11.84QMDD613 pKa = 3.9EE614 pKa = 4.36LDD616 pKa = 5.23LIDD619 pKa = 5.5AMTHH623 pKa = 4.67MRR625 pKa = 11.84IDD627 pKa = 3.78EE628 pKa = 4.44PDD630 pKa = 3.55KK631 pKa = 11.07LVQSLRR637 pKa = 11.84NTSRR641 pKa = 11.84SISHH645 pKa = 7.49IDD647 pKa = 3.52QLWKK651 pKa = 10.7KK652 pKa = 9.32GFKK655 pKa = 9.98RR656 pKa = 11.84LEE658 pKa = 4.35IINEE662 pKa = 3.83DD663 pKa = 3.4NYY665 pKa = 10.88VNFNEE670 pKa = 5.14LDD672 pKa = 3.78LPNKK676 pKa = 10.31NNLKK680 pKa = 9.79PWEE683 pKa = 4.22SYY685 pKa = 10.64YY686 pKa = 10.31ISNLSEE692 pKa = 4.74LSDD695 pKa = 3.97KK696 pKa = 11.0LDD698 pKa = 3.58SLKK701 pKa = 10.14TPVTQNMWFF710 pKa = 3.48
MM1 pKa = 6.92KK2 pKa = 10.18TKK4 pKa = 10.51FLVTKK9 pKa = 10.34RR10 pKa = 11.84LLEE13 pKa = 4.06SLFDD17 pKa = 4.36DD18 pKa = 3.52RR19 pKa = 11.84TIGKK23 pKa = 9.75KK24 pKa = 9.99YY25 pKa = 9.17IKK27 pKa = 10.22SVEE30 pKa = 3.95TMRR33 pKa = 11.84IKK35 pKa = 10.68SGLPFTIKK43 pKa = 10.49YY44 pKa = 7.28MKK46 pKa = 10.17AVKK49 pKa = 10.12LHH51 pKa = 4.48ITRR54 pKa = 11.84YY55 pKa = 9.89ISGQPLRR62 pKa = 11.84TNSSLVSLTNGFPTKK77 pKa = 10.49FLYY80 pKa = 10.5LKK82 pKa = 10.23EE83 pKa = 5.23LIDD86 pKa = 4.0SGDD89 pKa = 4.0PIKK92 pKa = 10.93LRR94 pKa = 11.84LVLTLTGYY102 pKa = 8.31TRR104 pKa = 11.84SIIPTKK110 pKa = 10.75DD111 pKa = 2.89EE112 pKa = 4.11EE113 pKa = 4.57KK114 pKa = 10.64LVKK117 pKa = 9.05PTFNSISDD125 pKa = 3.82PYY127 pKa = 10.16KK128 pKa = 10.22GKK130 pKa = 10.63EE131 pKa = 3.81NYY133 pKa = 7.81TIPTEE138 pKa = 4.53FINGFVEE145 pKa = 4.61KK146 pKa = 10.67YY147 pKa = 10.17KK148 pKa = 11.15LNFMPKK154 pKa = 8.37WDD156 pKa = 4.33NSLQYY161 pKa = 10.31ISNKK165 pKa = 9.71SSPFGKK171 pKa = 8.51STLTGPFALFHH182 pKa = 5.94MGHH185 pKa = 6.97WNLNMLDD192 pKa = 3.99HH193 pKa = 6.77FKK195 pKa = 11.33SLIGDD200 pKa = 4.12GSYY203 pKa = 10.93KK204 pKa = 10.54IMIGEE209 pKa = 4.02LLEE212 pKa = 4.99NVFKK216 pKa = 10.7DD217 pKa = 3.43HH218 pKa = 7.37RR219 pKa = 11.84CFHH222 pKa = 7.0IGNSLNGIGKK232 pKa = 9.06IAIVKK237 pKa = 10.29DD238 pKa = 3.58PEE240 pKa = 4.14LKK242 pKa = 10.01QRR244 pKa = 11.84AIAMVDD250 pKa = 3.61YY251 pKa = 10.72YY252 pKa = 11.06SQLVLRR258 pKa = 11.84PIHH261 pKa = 7.41DD262 pKa = 3.87GLLNKK267 pKa = 9.71LRR269 pKa = 11.84NLPQDD274 pKa = 3.11RR275 pKa = 11.84TFTQNPFNNWGKK287 pKa = 8.89TMGNSFWSLDD297 pKa = 3.57LSSATDD303 pKa = 3.75RR304 pKa = 11.84FPISLQEE311 pKa = 4.09KK312 pKa = 9.2VLASILKK319 pKa = 10.35DD320 pKa = 3.34EE321 pKa = 5.22AKK323 pKa = 10.82AEE325 pKa = 4.17TWRR328 pKa = 11.84KK329 pKa = 9.25ILIEE333 pKa = 4.31RR334 pKa = 11.84DD335 pKa = 3.38YY336 pKa = 11.49KK337 pKa = 11.09LPNGNLTRR345 pKa = 11.84YY346 pKa = 9.85SVGQPMGAYY355 pKa = 10.07SSWTAFTISHH365 pKa = 6.51HH366 pKa = 6.03LVVHH370 pKa = 6.49FAAHH374 pKa = 6.48LCGIEE379 pKa = 4.64NFDD382 pKa = 4.69KK383 pKa = 11.23YY384 pKa = 11.07ILLGDD389 pKa = 5.1DD390 pKa = 3.46IVINNDD396 pKa = 2.48KK397 pKa = 10.69VARR400 pKa = 11.84KK401 pKa = 9.27YY402 pKa = 10.71IKK404 pKa = 10.28IMTKK408 pKa = 10.23LGVDD412 pKa = 3.21ISINKK417 pKa = 7.45THH419 pKa = 5.9VSKK422 pKa = 9.84NTYY425 pKa = 9.41EE426 pKa = 4.06FAKK429 pKa = 10.2RR430 pKa = 11.84WVKK433 pKa = 10.74HH434 pKa = 5.65NIEE437 pKa = 3.86ITGIPLRR444 pKa = 11.84GILNNLNSLPTAIKK458 pKa = 9.31QLVYY462 pKa = 10.79YY463 pKa = 9.58FYY465 pKa = 11.34SCNTLWRR472 pKa = 11.84GNSVEE477 pKa = 4.75LIFSIFKK484 pKa = 10.15DD485 pKa = 2.82IKK487 pKa = 10.11LRR489 pKa = 11.84NRR491 pKa = 11.84FITDD495 pKa = 2.79SKK497 pKa = 11.3LRR499 pKa = 11.84ILSEE503 pKa = 3.74DD504 pKa = 3.64TIFVIRR510 pKa = 11.84QTLKK514 pKa = 10.12LNTYY518 pKa = 8.86EE519 pKa = 4.53EE520 pKa = 3.86IRR522 pKa = 11.84NYY524 pKa = 10.08FNKK527 pKa = 10.36KK528 pKa = 8.2LTIEE532 pKa = 4.08NVLIPNEE539 pKa = 4.01DD540 pKa = 3.62QIHH543 pKa = 5.4GFIRR547 pKa = 11.84VILCLGLNKK556 pKa = 9.82VAEE559 pKa = 4.25KK560 pKa = 10.72SGNDD564 pKa = 3.02LSKK567 pKa = 10.75YY568 pKa = 9.72YY569 pKa = 10.82KK570 pKa = 10.54SFEE573 pKa = 4.18KK574 pKa = 10.68NFRR577 pKa = 11.84KK578 pKa = 10.26GDD580 pKa = 3.59FDD582 pKa = 5.52LKK584 pKa = 10.38MLVRR588 pKa = 11.84HH589 pKa = 6.45PIVLGIYY596 pKa = 10.38NKK598 pKa = 9.63ISQMKK603 pKa = 8.8RR604 pKa = 11.84TLQKK608 pKa = 9.83VRR610 pKa = 11.84QMDD613 pKa = 3.9EE614 pKa = 4.36LDD616 pKa = 5.23LIDD619 pKa = 5.5AMTHH623 pKa = 4.67MRR625 pKa = 11.84IDD627 pKa = 3.78EE628 pKa = 4.44PDD630 pKa = 3.55KK631 pKa = 11.07LVQSLRR637 pKa = 11.84NTSRR641 pKa = 11.84SISHH645 pKa = 7.49IDD647 pKa = 3.52QLWKK651 pKa = 10.7KK652 pKa = 9.32GFKK655 pKa = 9.98RR656 pKa = 11.84LEE658 pKa = 4.35IINEE662 pKa = 3.83DD663 pKa = 3.4NYY665 pKa = 10.88VNFNEE670 pKa = 5.14LDD672 pKa = 3.78LPNKK676 pKa = 10.31NNLKK680 pKa = 9.79PWEE683 pKa = 4.22SYY685 pKa = 10.64YY686 pKa = 10.31ISNLSEE692 pKa = 4.74LSDD695 pKa = 3.97KK696 pKa = 11.0LDD698 pKa = 3.58SLKK701 pKa = 10.14TPVTQNMWFF710 pKa = 3.48
Molecular weight: 82.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
710 |
710 |
710 |
710.0 |
82.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.535 ± 0.0 | 0.563 ± 0.0 |
5.634 ± 0.0 | 4.648 ± 0.0 |
5.07 ± 0.0 | 4.93 ± 0.0 |
2.394 ± 0.0 | 9.014 ± 0.0 |
9.859 ± 0.0 | 11.549 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.394 ± 0.0 | 7.324 ± 0.0 |
3.662 ± 0.0 | 2.394 ± 0.0 |
4.789 ± 0.0 | 7.183 ± 0.0 |
5.915 ± 0.0 | 4.507 ± 0.0 |
1.549 ± 0.0 | 4.085 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
