
Shewanella woodyi (strain ATCC 51908 / MS32)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella; Shewanella woodyi
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
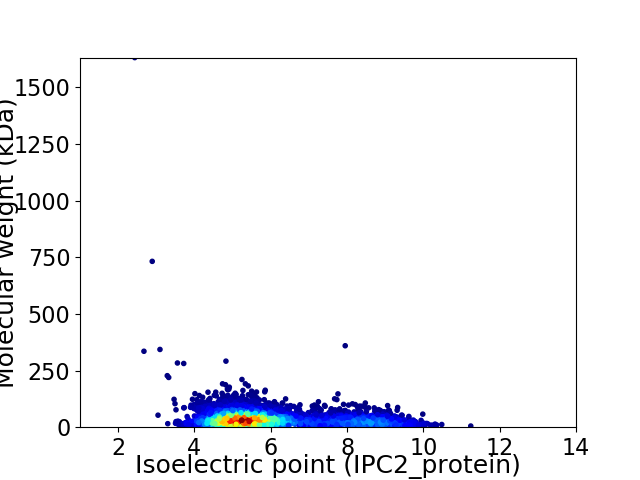
Virtual 2D-PAGE plot for 4865 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|B1KG88|GCSH_SHEWM Glycine cleavage system H protein OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=gcvH PE=3 SV=1
MM1 pKa = 7.28INKK4 pKa = 7.38VTFPLNCIALILASQLTGCGSDD26 pKa = 3.62GDD28 pKa = 4.52NDD30 pKa = 3.9SSTPTDD36 pKa = 3.38SAPPVITLVGNEE48 pKa = 4.31SIEE51 pKa = 4.11VVYY54 pKa = 10.96NAVFGDD60 pKa = 4.4LGAEE64 pKa = 4.01AHH66 pKa = 7.35DD67 pKa = 4.67DD68 pKa = 3.64TDD70 pKa = 4.05GSITVTTQGTVDD82 pKa = 3.05TSMPGTYY89 pKa = 9.85EE90 pKa = 3.64LTYY93 pKa = 10.4IATDD97 pKa = 3.27ASGNSSQATRR107 pKa = 11.84TVVVLEE113 pKa = 4.63DD114 pKa = 3.71TSQSATITTPGAPFSFFYY132 pKa = 11.12DD133 pKa = 3.8GVLKK137 pKa = 10.83GADD140 pKa = 3.48YY141 pKa = 10.12WSEE144 pKa = 3.93EE145 pKa = 3.95PQILSAGMGFDD156 pKa = 4.49GIVAIDD162 pKa = 3.93APEE165 pKa = 3.87LTIDD169 pKa = 3.67SVLAAGGAWSSDD181 pKa = 2.79IDD183 pKa = 4.52CGTGAQNYY191 pKa = 7.02TATSTRR197 pKa = 11.84DD198 pKa = 3.56QVSGAYY204 pKa = 9.5IIQTPYY210 pKa = 11.55GEE212 pKa = 4.97LKK214 pKa = 10.72DD215 pKa = 4.61DD216 pKa = 5.52AIGLDD221 pKa = 3.61GLPIVLSWPIDD232 pKa = 3.45TRR234 pKa = 11.84TLSLTDD240 pKa = 3.72FQFTLNTGDD249 pKa = 3.13IVRR252 pKa = 11.84PLAIGPLPNFEE263 pKa = 4.76DD264 pKa = 3.88NEE266 pKa = 4.51RR267 pKa = 11.84NTPVAFGEE275 pKa = 4.24FGNRR279 pKa = 11.84LPSDD283 pKa = 3.65HH284 pKa = 7.63PDD286 pKa = 2.61ARR288 pKa = 11.84FPIKK292 pKa = 10.78LEE294 pKa = 3.79IVEE297 pKa = 5.24DD298 pKa = 4.07DD299 pKa = 4.1TPLMMVGPGGQVVSAVGLTWEE320 pKa = 4.79TDD322 pKa = 3.06SSPYY326 pKa = 10.52DD327 pKa = 3.71EE328 pKa = 5.54NNGPRR333 pKa = 11.84LVGAKK338 pKa = 9.94LNRR341 pKa = 11.84IEE343 pKa = 4.09GQMDD347 pKa = 3.92GEE349 pKa = 5.14GISTLQPLIPANDD362 pKa = 3.31ATVLYY367 pKa = 10.79DD368 pKa = 3.94EE369 pKa = 5.2GDD371 pKa = 3.17FMLRR375 pKa = 11.84MLTSGGFSPDD385 pKa = 3.03GVSGLKK391 pKa = 10.23PNEE394 pKa = 3.6FEE396 pKa = 4.05RR397 pKa = 11.84FFRR400 pKa = 11.84IHH402 pKa = 7.35ANGLDD407 pKa = 3.78GNTVIIDD414 pKa = 3.77KK415 pKa = 10.93VGEE418 pKa = 4.07EE419 pKa = 4.44YY420 pKa = 10.77AVQGGTLRR428 pKa = 11.84VVGLSDD434 pKa = 4.53LGQPEE439 pKa = 4.1IGEE442 pKa = 4.43VTFDD446 pKa = 2.96ACYY449 pKa = 10.82SEE451 pKa = 6.1DD452 pKa = 3.62RR453 pKa = 11.84DD454 pKa = 3.81NYY456 pKa = 10.33IDD458 pKa = 4.51IILVGDD464 pKa = 3.79EE465 pKa = 3.66EE466 pKa = 4.25AARR469 pKa = 11.84NITILEE475 pKa = 4.16IPSLEE480 pKa = 4.03EE481 pKa = 4.56GYY483 pKa = 10.55SAFYY487 pKa = 10.93NPGGPGSTPFEE498 pKa = 4.24GVSYY502 pKa = 10.15SAPGPRR508 pKa = 11.84DD509 pKa = 3.65LEE511 pKa = 4.16PVIIALDD518 pKa = 3.5DD519 pKa = 3.99PMRR522 pKa = 11.84VTYY525 pKa = 10.7VPGSEE530 pKa = 4.13NVEE533 pKa = 4.08APDD536 pKa = 4.23NLMTFQFEE544 pKa = 4.62GIEE547 pKa = 4.21RR548 pKa = 11.84EE549 pKa = 4.02YY550 pKa = 11.21LLNVSSNYY558 pKa = 9.66TGDD561 pKa = 3.49VPVPLLFDD569 pKa = 3.43FHH571 pKa = 7.57GLNSAAEE578 pKa = 4.16VQYY581 pKa = 11.35SDD583 pKa = 3.91SQFNQISEE591 pKa = 4.44TEE593 pKa = 4.02NFILVTPQAINGWNVTGFPLGGNANDD619 pKa = 4.84LGFIDD624 pKa = 5.59ALIAQLSTAYY634 pKa = 10.82NIDD637 pKa = 3.36TNRR640 pKa = 11.84IYY642 pKa = 10.99AAGFSLGGFFSFEE655 pKa = 4.08LACQYY660 pKa = 11.38SDD662 pKa = 3.16TFAAIAPVSGVMTPAMAADD681 pKa = 4.57CVPEE685 pKa = 4.19RR686 pKa = 11.84PIPILQTHH694 pKa = 5.99GTADD698 pKa = 3.56DD699 pKa = 3.78QLPYY703 pKa = 10.92AQAQTVLQWWINFNQTDD720 pKa = 4.86LEE722 pKa = 4.2PVITDD727 pKa = 3.73LEE729 pKa = 4.4DD730 pKa = 3.61RR731 pKa = 11.84FPEE734 pKa = 4.32NGTTVQRR741 pKa = 11.84YY742 pKa = 8.43VYY744 pKa = 10.68GNGDD748 pKa = 3.27NGVSVEE754 pKa = 3.96HH755 pKa = 6.33LRR757 pKa = 11.84IEE759 pKa = 4.74GGQHH763 pKa = 5.6IWPGSAGDD771 pKa = 3.7SDD773 pKa = 4.18TNIAEE778 pKa = 4.63EE779 pKa = 3.59IWSFFEE785 pKa = 5.68SYY787 pKa = 11.11DD788 pKa = 4.03LNGKK792 pKa = 9.48INDD795 pKa = 3.64
MM1 pKa = 7.28INKK4 pKa = 7.38VTFPLNCIALILASQLTGCGSDD26 pKa = 3.62GDD28 pKa = 4.52NDD30 pKa = 3.9SSTPTDD36 pKa = 3.38SAPPVITLVGNEE48 pKa = 4.31SIEE51 pKa = 4.11VVYY54 pKa = 10.96NAVFGDD60 pKa = 4.4LGAEE64 pKa = 4.01AHH66 pKa = 7.35DD67 pKa = 4.67DD68 pKa = 3.64TDD70 pKa = 4.05GSITVTTQGTVDD82 pKa = 3.05TSMPGTYY89 pKa = 9.85EE90 pKa = 3.64LTYY93 pKa = 10.4IATDD97 pKa = 3.27ASGNSSQATRR107 pKa = 11.84TVVVLEE113 pKa = 4.63DD114 pKa = 3.71TSQSATITTPGAPFSFFYY132 pKa = 11.12DD133 pKa = 3.8GVLKK137 pKa = 10.83GADD140 pKa = 3.48YY141 pKa = 10.12WSEE144 pKa = 3.93EE145 pKa = 3.95PQILSAGMGFDD156 pKa = 4.49GIVAIDD162 pKa = 3.93APEE165 pKa = 3.87LTIDD169 pKa = 3.67SVLAAGGAWSSDD181 pKa = 2.79IDD183 pKa = 4.52CGTGAQNYY191 pKa = 7.02TATSTRR197 pKa = 11.84DD198 pKa = 3.56QVSGAYY204 pKa = 9.5IIQTPYY210 pKa = 11.55GEE212 pKa = 4.97LKK214 pKa = 10.72DD215 pKa = 4.61DD216 pKa = 5.52AIGLDD221 pKa = 3.61GLPIVLSWPIDD232 pKa = 3.45TRR234 pKa = 11.84TLSLTDD240 pKa = 3.72FQFTLNTGDD249 pKa = 3.13IVRR252 pKa = 11.84PLAIGPLPNFEE263 pKa = 4.76DD264 pKa = 3.88NEE266 pKa = 4.51RR267 pKa = 11.84NTPVAFGEE275 pKa = 4.24FGNRR279 pKa = 11.84LPSDD283 pKa = 3.65HH284 pKa = 7.63PDD286 pKa = 2.61ARR288 pKa = 11.84FPIKK292 pKa = 10.78LEE294 pKa = 3.79IVEE297 pKa = 5.24DD298 pKa = 4.07DD299 pKa = 4.1TPLMMVGPGGQVVSAVGLTWEE320 pKa = 4.79TDD322 pKa = 3.06SSPYY326 pKa = 10.52DD327 pKa = 3.71EE328 pKa = 5.54NNGPRR333 pKa = 11.84LVGAKK338 pKa = 9.94LNRR341 pKa = 11.84IEE343 pKa = 4.09GQMDD347 pKa = 3.92GEE349 pKa = 5.14GISTLQPLIPANDD362 pKa = 3.31ATVLYY367 pKa = 10.79DD368 pKa = 3.94EE369 pKa = 5.2GDD371 pKa = 3.17FMLRR375 pKa = 11.84MLTSGGFSPDD385 pKa = 3.03GVSGLKK391 pKa = 10.23PNEE394 pKa = 3.6FEE396 pKa = 4.05RR397 pKa = 11.84FFRR400 pKa = 11.84IHH402 pKa = 7.35ANGLDD407 pKa = 3.78GNTVIIDD414 pKa = 3.77KK415 pKa = 10.93VGEE418 pKa = 4.07EE419 pKa = 4.44YY420 pKa = 10.77AVQGGTLRR428 pKa = 11.84VVGLSDD434 pKa = 4.53LGQPEE439 pKa = 4.1IGEE442 pKa = 4.43VTFDD446 pKa = 2.96ACYY449 pKa = 10.82SEE451 pKa = 6.1DD452 pKa = 3.62RR453 pKa = 11.84DD454 pKa = 3.81NYY456 pKa = 10.33IDD458 pKa = 4.51IILVGDD464 pKa = 3.79EE465 pKa = 3.66EE466 pKa = 4.25AARR469 pKa = 11.84NITILEE475 pKa = 4.16IPSLEE480 pKa = 4.03EE481 pKa = 4.56GYY483 pKa = 10.55SAFYY487 pKa = 10.93NPGGPGSTPFEE498 pKa = 4.24GVSYY502 pKa = 10.15SAPGPRR508 pKa = 11.84DD509 pKa = 3.65LEE511 pKa = 4.16PVIIALDD518 pKa = 3.5DD519 pKa = 3.99PMRR522 pKa = 11.84VTYY525 pKa = 10.7VPGSEE530 pKa = 4.13NVEE533 pKa = 4.08APDD536 pKa = 4.23NLMTFQFEE544 pKa = 4.62GIEE547 pKa = 4.21RR548 pKa = 11.84EE549 pKa = 4.02YY550 pKa = 11.21LLNVSSNYY558 pKa = 9.66TGDD561 pKa = 3.49VPVPLLFDD569 pKa = 3.43FHH571 pKa = 7.57GLNSAAEE578 pKa = 4.16VQYY581 pKa = 11.35SDD583 pKa = 3.91SQFNQISEE591 pKa = 4.44TEE593 pKa = 4.02NFILVTPQAINGWNVTGFPLGGNANDD619 pKa = 4.84LGFIDD624 pKa = 5.59ALIAQLSTAYY634 pKa = 10.82NIDD637 pKa = 3.36TNRR640 pKa = 11.84IYY642 pKa = 10.99AAGFSLGGFFSFEE655 pKa = 4.08LACQYY660 pKa = 11.38SDD662 pKa = 3.16TFAAIAPVSGVMTPAMAADD681 pKa = 4.57CVPEE685 pKa = 4.19RR686 pKa = 11.84PIPILQTHH694 pKa = 5.99GTADD698 pKa = 3.56DD699 pKa = 3.78QLPYY703 pKa = 10.92AQAQTVLQWWINFNQTDD720 pKa = 4.86LEE722 pKa = 4.2PVITDD727 pKa = 3.73LEE729 pKa = 4.4DD730 pKa = 3.61RR731 pKa = 11.84FPEE734 pKa = 4.32NGTTVQRR741 pKa = 11.84YY742 pKa = 8.43VYY744 pKa = 10.68GNGDD748 pKa = 3.27NGVSVEE754 pKa = 3.96HH755 pKa = 6.33LRR757 pKa = 11.84IEE759 pKa = 4.74GGQHH763 pKa = 5.6IWPGSAGDD771 pKa = 3.7SDD773 pKa = 4.18TNIAEE778 pKa = 4.63EE779 pKa = 3.59IWSFFEE785 pKa = 5.68SYY787 pKa = 11.11DD788 pKa = 4.03LNGKK792 pKa = 9.48INDD795 pKa = 3.64
Molecular weight: 85.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B1KQ77|B1KQ77_SHEWM Uncharacterized protein OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=Swoo_0434 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPSTLKK12 pKa = 10.45RR13 pKa = 11.84KK14 pKa = 9.34RR15 pKa = 11.84SHH17 pKa = 6.2GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVSGRR29 pKa = 11.84KK30 pKa = 8.84VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.39GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPSTLKK12 pKa = 10.45RR13 pKa = 11.84KK14 pKa = 9.34RR15 pKa = 11.84SHH17 pKa = 6.2GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVSGRR29 pKa = 11.84KK30 pKa = 8.84VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.39GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1675288 |
32 |
16322 |
344.4 |
38.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.369 ± 0.042 | 1.026 ± 0.016 |
5.67 ± 0.069 | 6.271 ± 0.042 |
4.135 ± 0.032 | 6.973 ± 0.059 |
2.151 ± 0.027 | 6.422 ± 0.032 |
5.29 ± 0.059 | 10.582 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.542 ± 0.024 | 4.365 ± 0.037 |
3.812 ± 0.022 | 4.43 ± 0.036 |
4.162 ± 0.048 | 7.262 ± 0.042 |
5.547 ± 0.106 | 6.683 ± 0.046 |
1.217 ± 0.017 | 3.091 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
