
Clostridium leptum CAG:27
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
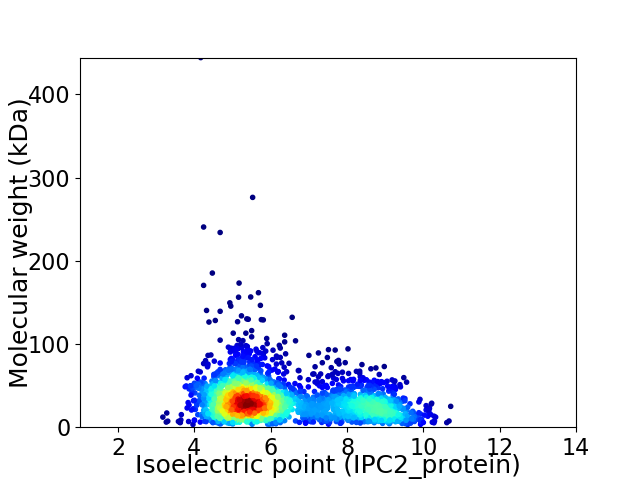
Virtual 2D-PAGE plot for 2482 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
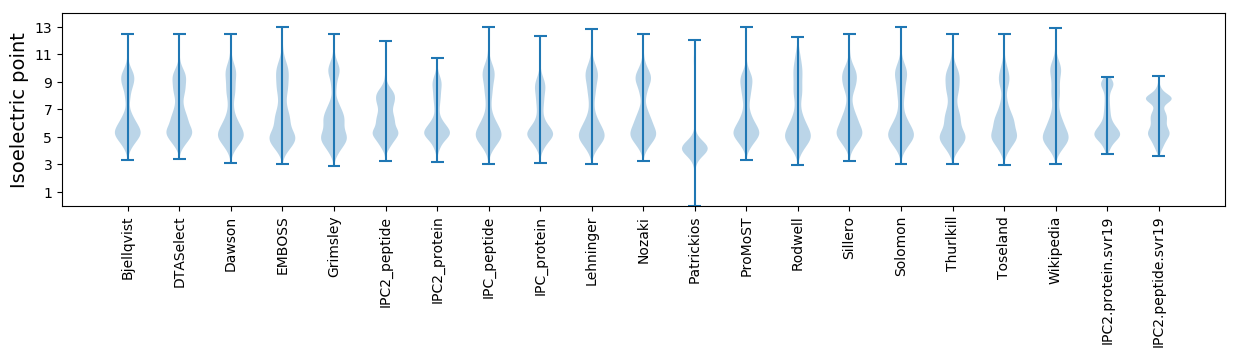
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6P1X5|R6P1X5_9CLOT Putative membrane protein OS=Clostridium leptum CAG:27 OX=1263068 GN=BN578_01953 PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 10.33KK3 pKa = 10.2VLALLLSGALLLSAAACSASPGGEE27 pKa = 3.73SSAGADD33 pKa = 3.16SSSTAEE39 pKa = 3.87EE40 pKa = 4.27SAAGASEE47 pKa = 4.41KK48 pKa = 10.27TSSTDD53 pKa = 3.11GQQLQTITFVLDD65 pKa = 3.13WTPNTNHH72 pKa = 6.24TGIYY76 pKa = 9.8VADD79 pKa = 3.47KK80 pKa = 10.87LGYY83 pKa = 9.65FKK85 pKa = 10.82EE86 pKa = 4.14AGIQIDD92 pKa = 3.63IQQPPEE98 pKa = 4.85DD99 pKa = 3.98GATALVATGKK109 pKa = 10.37AQFGVDD115 pKa = 3.97FQDD118 pKa = 3.84YY119 pKa = 9.59LAPAYY124 pKa = 10.86VSDD127 pKa = 4.43LPVTAVAALIQHH139 pKa = 5.51NTSGIISLKK148 pKa = 10.29EE149 pKa = 3.74KK150 pKa = 10.57GIQSPKK156 pKa = 10.13DD157 pKa = 3.68LEE159 pKa = 4.55GKK161 pKa = 8.45TYY163 pKa = 9.51ATWDD167 pKa = 3.89LPVEE171 pKa = 4.1QATMQNVVEE180 pKa = 4.38NDD182 pKa = 3.21GGDD185 pKa = 3.25WSKK188 pKa = 11.82VNLASVTVTDD198 pKa = 5.15VISALQTNIDD208 pKa = 4.04AVWIYY213 pKa = 8.68YY214 pKa = 10.21AWDD217 pKa = 4.96GIATQVKK224 pKa = 10.2GLDD227 pKa = 3.08TNYY230 pKa = 10.34FYY232 pKa = 10.96FKK234 pKa = 10.44DD235 pKa = 3.43INPVFDD241 pKa = 4.04YY242 pKa = 7.3YY243 pKa = 11.34TPVLVANNDD252 pKa = 3.61YY253 pKa = 11.27LEE255 pKa = 4.3QNPEE259 pKa = 3.67TAKK262 pKa = 11.05AFLAAVAKK270 pKa = 9.8GYY272 pKa = 10.58EE273 pKa = 4.02YY274 pKa = 10.55AIEE277 pKa = 4.31NPEE280 pKa = 3.69EE281 pKa = 3.87AAEE284 pKa = 4.2ILCEE288 pKa = 4.46ADD290 pKa = 3.13PTLDD294 pKa = 3.65SEE296 pKa = 4.96IVLSSQQWLADD307 pKa = 3.49QYY309 pKa = 11.07KK310 pKa = 10.71AEE312 pKa = 4.1VDD314 pKa = 2.59RR315 pKa = 11.84WGYY318 pKa = 10.32IDD320 pKa = 4.0PSRR323 pKa = 11.84WDD325 pKa = 4.88AFYY328 pKa = 10.78QWLWEE333 pKa = 3.96NQLIEE338 pKa = 4.24EE339 pKa = 5.08EE340 pKa = 4.32IPAGFGFSNDD350 pKa = 3.82YY351 pKa = 10.93LPEE354 pKa = 4.04
MM1 pKa = 7.38KK2 pKa = 10.33KK3 pKa = 10.2VLALLLSGALLLSAAACSASPGGEE27 pKa = 3.73SSAGADD33 pKa = 3.16SSSTAEE39 pKa = 3.87EE40 pKa = 4.27SAAGASEE47 pKa = 4.41KK48 pKa = 10.27TSSTDD53 pKa = 3.11GQQLQTITFVLDD65 pKa = 3.13WTPNTNHH72 pKa = 6.24TGIYY76 pKa = 9.8VADD79 pKa = 3.47KK80 pKa = 10.87LGYY83 pKa = 9.65FKK85 pKa = 10.82EE86 pKa = 4.14AGIQIDD92 pKa = 3.63IQQPPEE98 pKa = 4.85DD99 pKa = 3.98GATALVATGKK109 pKa = 10.37AQFGVDD115 pKa = 3.97FQDD118 pKa = 3.84YY119 pKa = 9.59LAPAYY124 pKa = 10.86VSDD127 pKa = 4.43LPVTAVAALIQHH139 pKa = 5.51NTSGIISLKK148 pKa = 10.29EE149 pKa = 3.74KK150 pKa = 10.57GIQSPKK156 pKa = 10.13DD157 pKa = 3.68LEE159 pKa = 4.55GKK161 pKa = 8.45TYY163 pKa = 9.51ATWDD167 pKa = 3.89LPVEE171 pKa = 4.1QATMQNVVEE180 pKa = 4.38NDD182 pKa = 3.21GGDD185 pKa = 3.25WSKK188 pKa = 11.82VNLASVTVTDD198 pKa = 5.15VISALQTNIDD208 pKa = 4.04AVWIYY213 pKa = 8.68YY214 pKa = 10.21AWDD217 pKa = 4.96GIATQVKK224 pKa = 10.2GLDD227 pKa = 3.08TNYY230 pKa = 10.34FYY232 pKa = 10.96FKK234 pKa = 10.44DD235 pKa = 3.43INPVFDD241 pKa = 4.04YY242 pKa = 7.3YY243 pKa = 11.34TPVLVANNDD252 pKa = 3.61YY253 pKa = 11.27LEE255 pKa = 4.3QNPEE259 pKa = 3.67TAKK262 pKa = 11.05AFLAAVAKK270 pKa = 9.8GYY272 pKa = 10.58EE273 pKa = 4.02YY274 pKa = 10.55AIEE277 pKa = 4.31NPEE280 pKa = 3.69EE281 pKa = 3.87AAEE284 pKa = 4.2ILCEE288 pKa = 4.46ADD290 pKa = 3.13PTLDD294 pKa = 3.65SEE296 pKa = 4.96IVLSSQQWLADD307 pKa = 3.49QYY309 pKa = 11.07KK310 pKa = 10.71AEE312 pKa = 4.1VDD314 pKa = 2.59RR315 pKa = 11.84WGYY318 pKa = 10.32IDD320 pKa = 4.0PSRR323 pKa = 11.84WDD325 pKa = 4.88AFYY328 pKa = 10.78QWLWEE333 pKa = 3.96NQLIEE338 pKa = 4.24EE339 pKa = 5.08EE340 pKa = 4.32IPAGFGFSNDD350 pKa = 3.82YY351 pKa = 10.93LPEE354 pKa = 4.04
Molecular weight: 38.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6NCX2|R6NCX2_9CLOT GTPase Der OS=Clostridium leptum CAG:27 OX=1263068 GN=der PE=3 SV=1
MM1 pKa = 7.63VSQQSKK7 pKa = 8.52TIRR10 pKa = 11.84NGKK13 pKa = 8.84KK14 pKa = 8.55EE15 pKa = 3.93RR16 pKa = 11.84TEE18 pKa = 3.29IFYY21 pKa = 10.61INNFIQTDD29 pKa = 4.38RR30 pKa = 11.84IKK32 pKa = 10.95SGSATSQFLSLAINITIFVLLSTYY56 pKa = 7.68SHH58 pKa = 6.28EE59 pKa = 4.27RR60 pKa = 11.84SFLIGYY66 pKa = 8.79RR67 pKa = 11.84RR68 pKa = 11.84AVVRR72 pKa = 11.84NCSPKK77 pKa = 10.65LLGRR81 pKa = 11.84LIRR84 pKa = 11.84KK85 pKa = 8.46GNQKK89 pKa = 9.76QDD91 pKa = 2.75SCFTVIGGRR100 pKa = 11.84SIFPIRR106 pKa = 11.84LCVYY110 pKa = 8.93WRR112 pKa = 11.84HH113 pKa = 6.43LEE115 pKa = 4.04LNNLFPEE122 pKa = 5.1PGDD125 pKa = 3.53PMIMLFQKK133 pKa = 10.69HH134 pKa = 5.67SFPFLKK140 pKa = 10.19RR141 pKa = 11.84RR142 pKa = 11.84NCIAGITNLKK152 pKa = 9.82
MM1 pKa = 7.63VSQQSKK7 pKa = 8.52TIRR10 pKa = 11.84NGKK13 pKa = 8.84KK14 pKa = 8.55EE15 pKa = 3.93RR16 pKa = 11.84TEE18 pKa = 3.29IFYY21 pKa = 10.61INNFIQTDD29 pKa = 4.38RR30 pKa = 11.84IKK32 pKa = 10.95SGSATSQFLSLAINITIFVLLSTYY56 pKa = 7.68SHH58 pKa = 6.28EE59 pKa = 4.27RR60 pKa = 11.84SFLIGYY66 pKa = 8.79RR67 pKa = 11.84RR68 pKa = 11.84AVVRR72 pKa = 11.84NCSPKK77 pKa = 10.65LLGRR81 pKa = 11.84LIRR84 pKa = 11.84KK85 pKa = 8.46GNQKK89 pKa = 9.76QDD91 pKa = 2.75SCFTVIGGRR100 pKa = 11.84SIFPIRR106 pKa = 11.84LCVYY110 pKa = 8.93WRR112 pKa = 11.84HH113 pKa = 6.43LEE115 pKa = 4.04LNNLFPEE122 pKa = 5.1PGDD125 pKa = 3.53PMIMLFQKK133 pKa = 10.69HH134 pKa = 5.67SFPFLKK140 pKa = 10.19RR141 pKa = 11.84RR142 pKa = 11.84NCIAGITNLKK152 pKa = 9.82
Molecular weight: 17.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
772695 |
29 |
4089 |
311.3 |
34.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.402 ± 0.049 | 1.627 ± 0.026 |
5.264 ± 0.044 | 7.082 ± 0.053 |
4.241 ± 0.044 | 7.581 ± 0.051 |
1.677 ± 0.022 | 6.439 ± 0.048 |
5.852 ± 0.039 | 9.553 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.811 ± 0.025 | 3.821 ± 0.035 |
3.972 ± 0.03 | 3.657 ± 0.027 |
4.942 ± 0.052 | 6.141 ± 0.046 |
5.123 ± 0.053 | 7.012 ± 0.037 |
1.038 ± 0.02 | 3.762 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
