
Sodalis-like symbiont of Bactericera trigonica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Bruguierivoracaceae; Sodalis; unclassified Sodalis
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
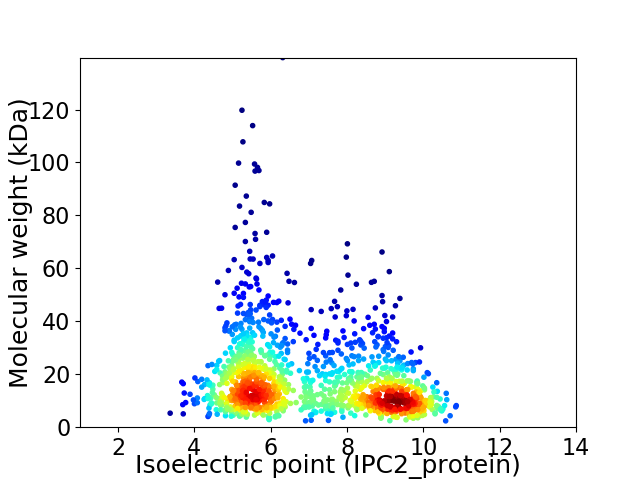
Virtual 2D-PAGE plot for 1308 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L7LSW5|A0A3L7LSW5_9GAMM Tail fiber domain-containing protein OS=Sodalis-like symbiont of Bactericera trigonica OX=2420513 GN=D8L93_03515 PE=4 SV=1
MM1 pKa = 7.59SEE3 pKa = 3.98QNNTEE8 pKa = 3.55IAFQIQRR15 pKa = 11.84IYY17 pKa = 10.13TKK19 pKa = 10.3DD20 pKa = 3.43VSFEE24 pKa = 4.02APNAPKK30 pKa = 10.45VFQQEE35 pKa = 3.95WQPEE39 pKa = 4.07IKK41 pKa = 10.36LDD43 pKa = 3.84LDD45 pKa = 3.92TASSQLAEE53 pKa = 5.2DD54 pKa = 3.5IYY56 pKa = 11.2EE57 pKa = 4.09VVLRR61 pKa = 11.84VTVTATLGEE70 pKa = 4.28DD71 pKa = 3.3TAFLCEE77 pKa = 4.07VQQAGIFTISGIDD90 pKa = 3.54STQMAHH96 pKa = 7.17CLGAYY101 pKa = 9.19CPNILFSYY109 pKa = 10.31ARR111 pKa = 11.84EE112 pKa = 4.45CITSQVSRR120 pKa = 11.84GTFPQLNLAPVNFDD134 pKa = 3.79TLFMNYY140 pKa = 9.3LQQQAAGVGSQPXQDD155 pKa = 4.1DD156 pKa = 3.37
MM1 pKa = 7.59SEE3 pKa = 3.98QNNTEE8 pKa = 3.55IAFQIQRR15 pKa = 11.84IYY17 pKa = 10.13TKK19 pKa = 10.3DD20 pKa = 3.43VSFEE24 pKa = 4.02APNAPKK30 pKa = 10.45VFQQEE35 pKa = 3.95WQPEE39 pKa = 4.07IKK41 pKa = 10.36LDD43 pKa = 3.84LDD45 pKa = 3.92TASSQLAEE53 pKa = 5.2DD54 pKa = 3.5IYY56 pKa = 11.2EE57 pKa = 4.09VVLRR61 pKa = 11.84VTVTATLGEE70 pKa = 4.28DD71 pKa = 3.3TAFLCEE77 pKa = 4.07VQQAGIFTISGIDD90 pKa = 3.54STQMAHH96 pKa = 7.17CLGAYY101 pKa = 9.19CPNILFSYY109 pKa = 10.31ARR111 pKa = 11.84EE112 pKa = 4.45CITSQVSRR120 pKa = 11.84GTFPQLNLAPVNFDD134 pKa = 3.79TLFMNYY140 pKa = 9.3LQQQAAGVGSQPXQDD155 pKa = 4.1DD156 pKa = 3.37
Molecular weight: 17.24 kDa
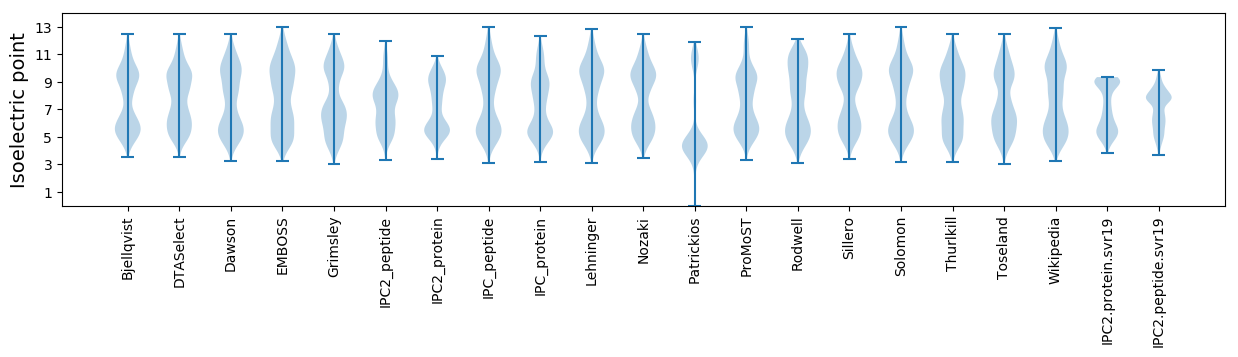
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L7LS07|A0A3L7LS07_9GAMM Imidazole glycerol phosphate synthase subunit HisH OS=Sodalis-like symbiont of Bactericera trigonica OX=2420513 GN=hisH PE=3 SV=1
MM1 pKa = 7.29ATQARR6 pKa = 11.84QSWPRR11 pKa = 11.84TSPTGDD17 pKa = 3.03AARR20 pKa = 11.84NNVCLFVANDD30 pKa = 4.07LEE32 pKa = 4.69YY33 pKa = 10.24PLPDD37 pKa = 2.99HH38 pKa = 6.53VFRR41 pKa = 11.84GTLLPTPLSPWCRR54 pKa = 11.84VANVRR59 pKa = 11.84QGVLVLEE66 pKa = 4.48TANASWKK73 pKa = 9.02MRR75 pKa = 11.84LRR77 pKa = 11.84YY78 pKa = 7.44EE79 pKa = 4.17QPRR82 pKa = 11.84LLSALRR88 pKa = 11.84AQILPSLSSIDD99 pKa = 3.36IRR101 pKa = 11.84INPSMARR108 pKa = 11.84KK109 pKa = 9.49QEE111 pKa = 4.16LNAQNKK117 pKa = 10.12DD118 pKa = 2.89SGQQLDD124 pKa = 4.15RR125 pKa = 11.84PPGEE129 pKa = 3.82KK130 pKa = 9.48PRR132 pKa = 11.84RR133 pKa = 11.84LSVQSAASIRR143 pKa = 11.84HH144 pKa = 3.98VAEE147 pKa = 3.98RR148 pKa = 11.84SEE150 pKa = 4.52GKK152 pKa = 10.12LKK154 pKa = 10.85NALEE158 pKa = 4.21RR159 pKa = 11.84LAEE162 pKa = 3.99LASRR166 pKa = 11.84RR167 pKa = 11.84EE168 pKa = 4.18CQPRR172 pKa = 11.84PP173 pKa = 3.39
MM1 pKa = 7.29ATQARR6 pKa = 11.84QSWPRR11 pKa = 11.84TSPTGDD17 pKa = 3.03AARR20 pKa = 11.84NNVCLFVANDD30 pKa = 4.07LEE32 pKa = 4.69YY33 pKa = 10.24PLPDD37 pKa = 2.99HH38 pKa = 6.53VFRR41 pKa = 11.84GTLLPTPLSPWCRR54 pKa = 11.84VANVRR59 pKa = 11.84QGVLVLEE66 pKa = 4.48TANASWKK73 pKa = 9.02MRR75 pKa = 11.84LRR77 pKa = 11.84YY78 pKa = 7.44EE79 pKa = 4.17QPRR82 pKa = 11.84LLSALRR88 pKa = 11.84AQILPSLSSIDD99 pKa = 3.36IRR101 pKa = 11.84INPSMARR108 pKa = 11.84KK109 pKa = 9.49QEE111 pKa = 4.16LNAQNKK117 pKa = 10.12DD118 pKa = 2.89SGQQLDD124 pKa = 4.15RR125 pKa = 11.84PPGEE129 pKa = 3.82KK130 pKa = 9.48PRR132 pKa = 11.84RR133 pKa = 11.84LSVQSAASIRR143 pKa = 11.84HH144 pKa = 3.98VAEE147 pKa = 3.98RR148 pKa = 11.84SEE150 pKa = 4.52GKK152 pKa = 10.12LKK154 pKa = 10.85NALEE158 pKa = 4.21RR159 pKa = 11.84LAEE162 pKa = 3.99LASRR166 pKa = 11.84RR167 pKa = 11.84EE168 pKa = 4.18CQPRR172 pKa = 11.84PP173 pKa = 3.39
Molecular weight: 19.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
230015 |
21 |
1271 |
175.9 |
19.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.305 ± 0.096 | 1.244 ± 0.028 |
5.311 ± 0.053 | 5.441 ± 0.077 |
3.462 ± 0.05 | 7.386 ± 0.081 |
2.419 ± 0.038 | 5.485 ± 0.065 |
3.964 ± 0.072 | 11.135 ± 0.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.742 ± 0.042 | 3.358 ± 0.044 |
4.509 ± 0.048 | 4.606 ± 0.069 |
6.863 ± 0.067 | 5.623 ± 0.063 |
5.091 ± 0.058 | 6.956 ± 0.075 |
1.327 ± 0.037 | 2.739 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
