
bacterium HR39
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
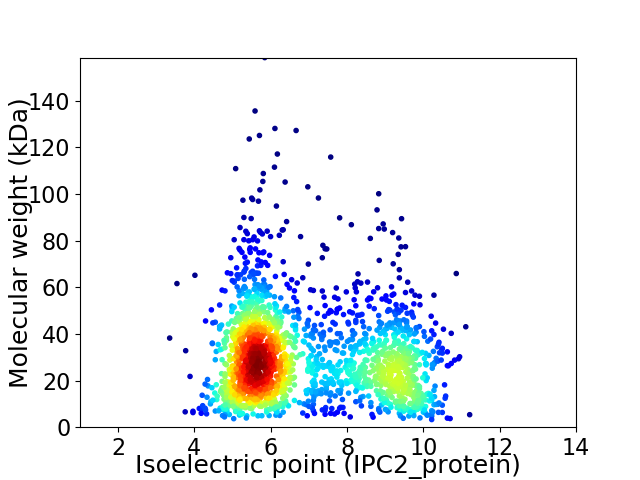
Virtual 2D-PAGE plot for 1916 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H6ATD8|A0A2H6ATD8_9BACT Superoxide dismutase OS=bacterium HR39 OX=2035434 GN=sodB PE=3 SV=1
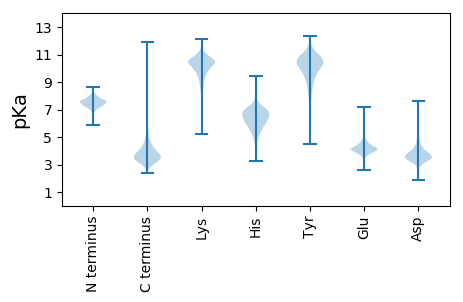
MM1 pKa = 7.57SEE3 pKa = 4.13RR4 pKa = 11.84SVDD7 pKa = 2.99RR8 pKa = 11.84WIEE11 pKa = 3.95VATLDD16 pKa = 5.1DD17 pKa = 3.69LWEE20 pKa = 4.64GDD22 pKa = 3.84FLDD25 pKa = 4.5VEE27 pKa = 4.61VDD29 pKa = 3.55GEE31 pKa = 4.79TVLLVHH37 pKa = 6.66LPGGAIRR44 pKa = 11.84AYY46 pKa = 10.46QGICPHH52 pKa = 5.79QEE54 pKa = 3.05IALADD59 pKa = 3.59GDD61 pKa = 4.35FDD63 pKa = 4.45FEE65 pKa = 5.37SGILTCSAHH74 pKa = 6.02RR75 pKa = 11.84WQFEE79 pKa = 3.97LATGQGVNPADD90 pKa = 3.53CWLYY94 pKa = 9.38EE95 pKa = 4.1YY96 pKa = 9.89PVKK99 pKa = 11.13VEE101 pKa = 4.28DD102 pKa = 3.81DD103 pKa = 3.72KK104 pKa = 11.71IYY106 pKa = 10.45IGWVEE111 pKa = 4.13GEE113 pKa = 3.79RR114 pKa = 11.84HH115 pKa = 5.17YY116 pKa = 11.38NRR118 pKa = 11.84CAGG121 pKa = 3.5
MM1 pKa = 7.57SEE3 pKa = 4.13RR4 pKa = 11.84SVDD7 pKa = 2.99RR8 pKa = 11.84WIEE11 pKa = 3.95VATLDD16 pKa = 5.1DD17 pKa = 3.69LWEE20 pKa = 4.64GDD22 pKa = 3.84FLDD25 pKa = 4.5VEE27 pKa = 4.61VDD29 pKa = 3.55GEE31 pKa = 4.79TVLLVHH37 pKa = 6.66LPGGAIRR44 pKa = 11.84AYY46 pKa = 10.46QGICPHH52 pKa = 5.79QEE54 pKa = 3.05IALADD59 pKa = 3.59GDD61 pKa = 4.35FDD63 pKa = 4.45FEE65 pKa = 5.37SGILTCSAHH74 pKa = 6.02RR75 pKa = 11.84WQFEE79 pKa = 3.97LATGQGVNPADD90 pKa = 3.53CWLYY94 pKa = 9.38EE95 pKa = 4.1YY96 pKa = 9.89PVKK99 pKa = 11.13VEE101 pKa = 4.28DD102 pKa = 3.81DD103 pKa = 3.72KK104 pKa = 11.71IYY106 pKa = 10.45IGWVEE111 pKa = 4.13GEE113 pKa = 3.79RR114 pKa = 11.84HH115 pKa = 5.17YY116 pKa = 11.38NRR118 pKa = 11.84CAGG121 pKa = 3.5
Molecular weight: 13.69 kDa
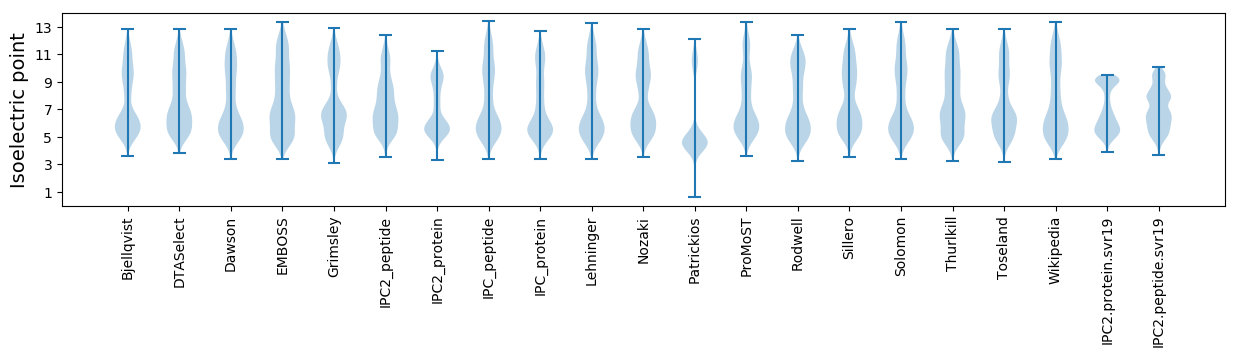
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H6AS14|A0A2H6AS14_9BACT EVE domain-containing protein OS=bacterium HR39 OX=2035434 GN=HRbin39_00084 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84PRR4 pKa = 11.84TGPSSRR10 pKa = 11.84PRR12 pKa = 11.84RR13 pKa = 11.84LLGTSRR19 pKa = 11.84LRR21 pKa = 11.84AGRR24 pKa = 11.84PASGPARR31 pKa = 11.84LPARR35 pKa = 11.84LSRR38 pKa = 11.84LRR40 pKa = 11.84PHH42 pKa = 6.17GWGRR46 pKa = 11.84TGGSPGRR53 pKa = 11.84SAPRR57 pKa = 11.84HH58 pKa = 5.08RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84LAADD65 pKa = 3.78LRR67 pKa = 11.84ARR69 pKa = 11.84GGGGPGGSRR78 pKa = 11.84GRR80 pKa = 11.84APLLHH85 pKa = 6.81RR86 pKa = 11.84ALPPARR92 pKa = 11.84RR93 pKa = 11.84GRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84GRR99 pKa = 11.84PRR101 pKa = 11.84ALHH104 pKa = 5.83RR105 pKa = 11.84LLRR108 pKa = 11.84TGALGLARR116 pKa = 11.84ARR118 pKa = 11.84RR119 pKa = 11.84PLRR122 pKa = 11.84PSPLRR127 pKa = 11.84PPAGSGGRR135 pKa = 11.84GPRR138 pKa = 11.84PVRR141 pKa = 11.84PRR143 pKa = 11.84AARR146 pKa = 11.84PAHH149 pKa = 5.9RR150 pKa = 11.84RR151 pKa = 11.84AGRR154 pKa = 11.84GHH156 pKa = 7.47AARR159 pKa = 11.84SLLRR163 pKa = 11.84PRR165 pKa = 11.84RR166 pKa = 11.84HH167 pKa = 5.31RR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84ARR173 pKa = 11.84RR174 pKa = 11.84ARR176 pKa = 11.84AGTAVGGRR184 pKa = 11.84SRR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84LLPPHH193 pKa = 6.75PGIGPGAPSRR203 pKa = 11.84RR204 pKa = 11.84TGAAGGLRR212 pKa = 11.84RR213 pKa = 11.84TEE215 pKa = 3.97RR216 pKa = 11.84PSLPRR221 pKa = 11.84HH222 pKa = 5.7RR223 pKa = 11.84PRR225 pKa = 11.84SRR227 pKa = 11.84GDD229 pKa = 2.96GRR231 pKa = 11.84ALARR235 pKa = 11.84GGLAVLHH242 pKa = 6.58PRR244 pKa = 11.84LAHH247 pKa = 5.76GPSRR251 pKa = 11.84PRR253 pKa = 11.84PRR255 pKa = 11.84AAAPQPLLRR264 pKa = 11.84LLPHH268 pKa = 6.72PRR270 pKa = 11.84RR271 pKa = 11.84GAWGRR276 pKa = 11.84GSARR280 pKa = 11.84SDD282 pKa = 2.91GCAAHH287 pKa = 7.12AGTFHH292 pKa = 7.56RR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84PPLLAARR302 pKa = 11.84RR303 pKa = 11.84ALLRR307 pKa = 11.84RR308 pKa = 11.84CSGPLPRR315 pKa = 11.84GRR317 pKa = 11.84TAVAPAGGGAGHH329 pKa = 7.08RR330 pKa = 11.84RR331 pKa = 11.84RR332 pKa = 11.84HH333 pKa = 5.36PRR335 pKa = 11.84APPRR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84LLGLGQAGRR350 pKa = 11.84IHH352 pKa = 6.43GRR354 pKa = 11.84PHH356 pKa = 6.05EE357 pKa = 4.29ARR359 pKa = 11.84GADD362 pKa = 3.5VGAGAARR369 pKa = 11.84GDD371 pKa = 3.43GHH373 pKa = 6.77RR374 pKa = 11.84RR375 pKa = 11.84GPRR378 pKa = 11.84GTTASGGGAPLLLPCAAAVRR398 pKa = 11.84TGEE401 pKa = 4.09GMPP404 pKa = 3.92
MM1 pKa = 7.97RR2 pKa = 11.84PRR4 pKa = 11.84TGPSSRR10 pKa = 11.84PRR12 pKa = 11.84RR13 pKa = 11.84LLGTSRR19 pKa = 11.84LRR21 pKa = 11.84AGRR24 pKa = 11.84PASGPARR31 pKa = 11.84LPARR35 pKa = 11.84LSRR38 pKa = 11.84LRR40 pKa = 11.84PHH42 pKa = 6.17GWGRR46 pKa = 11.84TGGSPGRR53 pKa = 11.84SAPRR57 pKa = 11.84HH58 pKa = 5.08RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84LAADD65 pKa = 3.78LRR67 pKa = 11.84ARR69 pKa = 11.84GGGGPGGSRR78 pKa = 11.84GRR80 pKa = 11.84APLLHH85 pKa = 6.81RR86 pKa = 11.84ALPPARR92 pKa = 11.84RR93 pKa = 11.84GRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84GRR99 pKa = 11.84PRR101 pKa = 11.84ALHH104 pKa = 5.83RR105 pKa = 11.84LLRR108 pKa = 11.84TGALGLARR116 pKa = 11.84ARR118 pKa = 11.84RR119 pKa = 11.84PLRR122 pKa = 11.84PSPLRR127 pKa = 11.84PPAGSGGRR135 pKa = 11.84GPRR138 pKa = 11.84PVRR141 pKa = 11.84PRR143 pKa = 11.84AARR146 pKa = 11.84PAHH149 pKa = 5.9RR150 pKa = 11.84RR151 pKa = 11.84AGRR154 pKa = 11.84GHH156 pKa = 7.47AARR159 pKa = 11.84SLLRR163 pKa = 11.84PRR165 pKa = 11.84RR166 pKa = 11.84HH167 pKa = 5.31RR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84ARR173 pKa = 11.84RR174 pKa = 11.84ARR176 pKa = 11.84AGTAVGGRR184 pKa = 11.84SRR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84LLPPHH193 pKa = 6.75PGIGPGAPSRR203 pKa = 11.84RR204 pKa = 11.84TGAAGGLRR212 pKa = 11.84RR213 pKa = 11.84TEE215 pKa = 3.97RR216 pKa = 11.84PSLPRR221 pKa = 11.84HH222 pKa = 5.7RR223 pKa = 11.84PRR225 pKa = 11.84SRR227 pKa = 11.84GDD229 pKa = 2.96GRR231 pKa = 11.84ALARR235 pKa = 11.84GGLAVLHH242 pKa = 6.58PRR244 pKa = 11.84LAHH247 pKa = 5.76GPSRR251 pKa = 11.84PRR253 pKa = 11.84PRR255 pKa = 11.84AAAPQPLLRR264 pKa = 11.84LLPHH268 pKa = 6.72PRR270 pKa = 11.84RR271 pKa = 11.84GAWGRR276 pKa = 11.84GSARR280 pKa = 11.84SDD282 pKa = 2.91GCAAHH287 pKa = 7.12AGTFHH292 pKa = 7.56RR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84PPLLAARR302 pKa = 11.84RR303 pKa = 11.84ALLRR307 pKa = 11.84RR308 pKa = 11.84CSGPLPRR315 pKa = 11.84GRR317 pKa = 11.84TAVAPAGGGAGHH329 pKa = 7.08RR330 pKa = 11.84RR331 pKa = 11.84RR332 pKa = 11.84HH333 pKa = 5.36PRR335 pKa = 11.84APPRR339 pKa = 11.84RR340 pKa = 11.84RR341 pKa = 11.84LLGLGQAGRR350 pKa = 11.84IHH352 pKa = 6.43GRR354 pKa = 11.84PHH356 pKa = 6.05EE357 pKa = 4.29ARR359 pKa = 11.84GADD362 pKa = 3.5VGAGAARR369 pKa = 11.84GDD371 pKa = 3.43GHH373 pKa = 6.77RR374 pKa = 11.84RR375 pKa = 11.84GPRR378 pKa = 11.84GTTASGGGAPLLLPCAAAVRR398 pKa = 11.84TGEE401 pKa = 4.09GMPP404 pKa = 3.92
Molecular weight: 43.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
551194 |
29 |
1434 |
287.7 |
31.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.234 ± 0.081 | 0.878 ± 0.02 |
5.048 ± 0.045 | 7.316 ± 0.062 |
3.302 ± 0.042 | 9.18 ± 0.062 |
2.337 ± 0.037 | 3.473 ± 0.046 |
1.648 ± 0.029 | 11.462 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.776 ± 0.027 | 1.392 ± 0.025 |
6.577 ± 0.049 | 2.234 ± 0.036 |
10.647 ± 0.076 | 3.598 ± 0.038 |
4.096 ± 0.034 | 8.676 ± 0.055 |
1.388 ± 0.025 | 1.738 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
