
Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus thermophilus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
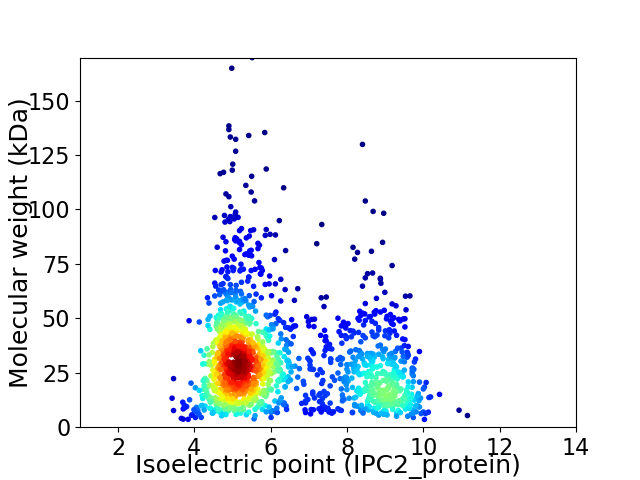
Virtual 2D-PAGE plot for 1577 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5M5M7|Y441_STRT2 UPF0346 protein stu0441 OS=Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) OX=264199 GN=stu0441 PE=3 SV=1
MM1 pKa = 7.52LSKK4 pKa = 10.94SKK6 pKa = 7.14TTKK9 pKa = 10.54ALLYY13 pKa = 9.35STAALSLFAASHH25 pKa = 4.36VHH27 pKa = 7.09ADD29 pKa = 3.94EE30 pKa = 4.49TSHH33 pKa = 4.16WTARR37 pKa = 11.84SVDD40 pKa = 3.79QIKK43 pKa = 11.06ADD45 pKa = 3.1ISVNDD50 pKa = 3.47NQQTYY55 pKa = 7.44TVQYY59 pKa = 10.1GDD61 pKa = 3.61TLGSIVEE68 pKa = 4.11AMGIDD73 pKa = 3.8MNVLANIKK81 pKa = 10.03EE82 pKa = 4.0ITNIDD87 pKa = 4.33LIFPGTVLTTTYY99 pKa = 10.87NADD102 pKa = 3.45NQAVSVKK109 pKa = 10.39VEE111 pKa = 4.34TPSSEE116 pKa = 4.19TSDD119 pKa = 3.54TPVVAEE125 pKa = 4.68SNLTTNEE132 pKa = 3.73VTVNGQSVVASDD144 pKa = 3.82LSAPVEE150 pKa = 4.56TVSLTATQAPAKK162 pKa = 9.83EE163 pKa = 4.44EE164 pKa = 4.06STQVVSEE171 pKa = 4.18VTEE174 pKa = 5.01AIASASDD181 pKa = 3.26TPAYY185 pKa = 10.45ADD187 pKa = 3.6TEE189 pKa = 4.45QPVADD194 pKa = 5.58AIDD197 pKa = 4.14HH198 pKa = 5.3VTSSAEE204 pKa = 3.61EE205 pKa = 3.83TLAEE209 pKa = 4.14EE210 pKa = 4.22EE211 pKa = 4.45APATEE216 pKa = 4.4TSAQAEE222 pKa = 4.48TTEE225 pKa = 4.18VAATSEE231 pKa = 4.33AASEE235 pKa = 4.17AASEE239 pKa = 4.58VPAEE243 pKa = 4.0QLAAASEE250 pKa = 4.33APEE253 pKa = 4.25SSEE256 pKa = 5.06APAEE260 pKa = 4.16QPAAAPEE267 pKa = 4.32SSEE270 pKa = 4.72APAEE274 pKa = 4.18QPAATSEE281 pKa = 4.22AASEE285 pKa = 4.42APASVVPVATSEE297 pKa = 4.29AVSEE301 pKa = 4.27APAVSEE307 pKa = 4.69VPAEE311 pKa = 3.97QLAAASEE318 pKa = 4.35APEE321 pKa = 4.19SSEE324 pKa = 4.69VPAEE328 pKa = 4.13QPAAAPEE335 pKa = 4.32SSEE338 pKa = 4.72APAEE342 pKa = 4.18QPAATSEE349 pKa = 4.11AAPATSEE356 pKa = 4.35APAEE360 pKa = 3.94QLAATSEE367 pKa = 4.25AASTPNTYY375 pKa = 10.0PVGQCTWGAKK385 pKa = 9.56SLAPWAGNNWGNAKK399 pKa = 10.27DD400 pKa = 4.0WIASAQAAGHH410 pKa = 5.73SVGTTPVAGAIAVWPNDD427 pKa = 3.19GGGYY431 pKa = 9.63GHH433 pKa = 6.72VAYY436 pKa = 8.98VTSASGVNSIQVMEE450 pKa = 4.5SNYY453 pKa = 10.27AGNMLIGNYY462 pKa = 9.71RR463 pKa = 11.84GTFDD467 pKa = 3.96PTSSAHH473 pKa = 6.21GGSVYY478 pKa = 10.67YY479 pKa = 10.14IYY481 pKa = 10.65PP482 pKa = 3.8
MM1 pKa = 7.52LSKK4 pKa = 10.94SKK6 pKa = 7.14TTKK9 pKa = 10.54ALLYY13 pKa = 9.35STAALSLFAASHH25 pKa = 4.36VHH27 pKa = 7.09ADD29 pKa = 3.94EE30 pKa = 4.49TSHH33 pKa = 4.16WTARR37 pKa = 11.84SVDD40 pKa = 3.79QIKK43 pKa = 11.06ADD45 pKa = 3.1ISVNDD50 pKa = 3.47NQQTYY55 pKa = 7.44TVQYY59 pKa = 10.1GDD61 pKa = 3.61TLGSIVEE68 pKa = 4.11AMGIDD73 pKa = 3.8MNVLANIKK81 pKa = 10.03EE82 pKa = 4.0ITNIDD87 pKa = 4.33LIFPGTVLTTTYY99 pKa = 10.87NADD102 pKa = 3.45NQAVSVKK109 pKa = 10.39VEE111 pKa = 4.34TPSSEE116 pKa = 4.19TSDD119 pKa = 3.54TPVVAEE125 pKa = 4.68SNLTTNEE132 pKa = 3.73VTVNGQSVVASDD144 pKa = 3.82LSAPVEE150 pKa = 4.56TVSLTATQAPAKK162 pKa = 9.83EE163 pKa = 4.44EE164 pKa = 4.06STQVVSEE171 pKa = 4.18VTEE174 pKa = 5.01AIASASDD181 pKa = 3.26TPAYY185 pKa = 10.45ADD187 pKa = 3.6TEE189 pKa = 4.45QPVADD194 pKa = 5.58AIDD197 pKa = 4.14HH198 pKa = 5.3VTSSAEE204 pKa = 3.61EE205 pKa = 3.83TLAEE209 pKa = 4.14EE210 pKa = 4.22EE211 pKa = 4.45APATEE216 pKa = 4.4TSAQAEE222 pKa = 4.48TTEE225 pKa = 4.18VAATSEE231 pKa = 4.33AASEE235 pKa = 4.17AASEE239 pKa = 4.58VPAEE243 pKa = 4.0QLAAASEE250 pKa = 4.33APEE253 pKa = 4.25SSEE256 pKa = 5.06APAEE260 pKa = 4.16QPAAAPEE267 pKa = 4.32SSEE270 pKa = 4.72APAEE274 pKa = 4.18QPAATSEE281 pKa = 4.22AASEE285 pKa = 4.42APASVVPVATSEE297 pKa = 4.29AVSEE301 pKa = 4.27APAVSEE307 pKa = 4.69VPAEE311 pKa = 3.97QLAAASEE318 pKa = 4.35APEE321 pKa = 4.19SSEE324 pKa = 4.69VPAEE328 pKa = 4.13QPAAAPEE335 pKa = 4.32SSEE338 pKa = 4.72APAEE342 pKa = 4.18QPAATSEE349 pKa = 4.11AAPATSEE356 pKa = 4.35APAEE360 pKa = 3.94QLAATSEE367 pKa = 4.25AASTPNTYY375 pKa = 10.0PVGQCTWGAKK385 pKa = 9.56SLAPWAGNNWGNAKK399 pKa = 10.27DD400 pKa = 4.0WIASAQAAGHH410 pKa = 5.73SVGTTPVAGAIAVWPNDD427 pKa = 3.19GGGYY431 pKa = 9.63GHH433 pKa = 6.72VAYY436 pKa = 8.98VTSASGVNSIQVMEE450 pKa = 4.5SNYY453 pKa = 10.27AGNMLIGNYY462 pKa = 9.71RR463 pKa = 11.84GTFDD467 pKa = 3.96PTSSAHH473 pKa = 6.21GGSVYY478 pKa = 10.67YY479 pKa = 10.14IYY481 pKa = 10.65PP482 pKa = 3.8
Molecular weight: 48.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5M2L3|Q5M2L3_STRT2 CoA_binding domain-containing protein OS=Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) OX=264199 GN=stu1802 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.67IRR11 pKa = 11.84RR12 pKa = 11.84QRR14 pKa = 11.84KK15 pKa = 7.52HH16 pKa = 5.2GFRR19 pKa = 11.84HH20 pKa = 6.39RR21 pKa = 11.84MSTKK25 pKa = 9.02NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.87GRR39 pKa = 11.84KK40 pKa = 8.75VLAAA44 pKa = 4.31
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.67IRR11 pKa = 11.84RR12 pKa = 11.84QRR14 pKa = 11.84KK15 pKa = 7.52HH16 pKa = 5.2GFRR19 pKa = 11.84HH20 pKa = 6.39RR21 pKa = 11.84MSTKK25 pKa = 9.02NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.87GRR39 pKa = 11.84KK40 pKa = 8.75VLAAA44 pKa = 4.31
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
458119 |
31 |
1470 |
290.5 |
32.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.484 ± 0.073 | 0.563 ± 0.016 |
5.87 ± 0.056 | 6.868 ± 0.068 |
4.607 ± 0.042 | 6.597 ± 0.058 |
1.893 ± 0.027 | 7.431 ± 0.059 |
6.879 ± 0.066 | 9.965 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.539 ± 0.03 | 4.59 ± 0.04 |
3.251 ± 0.033 | 3.732 ± 0.043 |
4.12 ± 0.048 | 6.239 ± 0.061 |
5.67 ± 0.047 | 7.15 ± 0.054 |
0.831 ± 0.02 | 3.723 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
