
Variovorax paradoxus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Variovorax
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
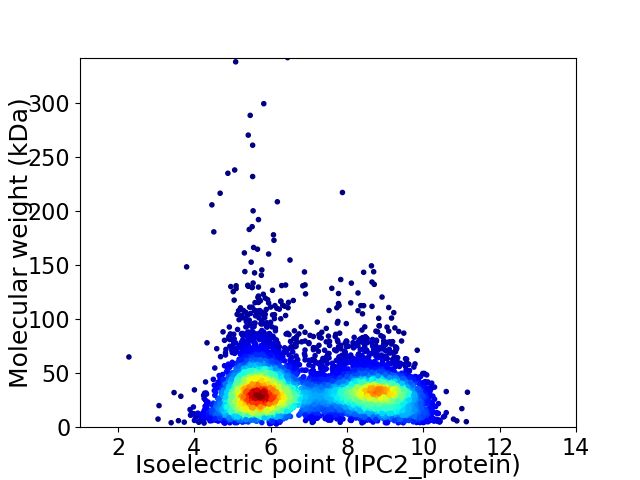
Virtual 2D-PAGE plot for 6788 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H2LW41|A0A0H2LW41_VARPD Multidrug resistance protein MdtB OS=Variovorax paradoxus OX=34073 GN=mdtB PE=4 SV=1
MM1 pKa = 6.94EE2 pKa = 5.37AVYY5 pKa = 10.6KK6 pKa = 10.85SGGASRR12 pKa = 11.84KK13 pKa = 9.22AAADD17 pKa = 3.66TPLVLDD23 pKa = 4.13RR24 pKa = 11.84PSIVSLKK31 pKa = 10.04IAPEE35 pKa = 3.99SVLKK39 pKa = 9.89FEE41 pKa = 4.98RR42 pKa = 11.84RR43 pKa = 11.84GGDD46 pKa = 3.25LVLVLQGGQEE56 pKa = 3.97VAVRR60 pKa = 11.84GFFTEE65 pKa = 4.1YY66 pKa = 10.84SDD68 pKa = 4.27GGRR71 pKa = 11.84NDD73 pKa = 4.32LVLEE77 pKa = 4.93DD78 pKa = 4.05GAGVQWWGQYY88 pKa = 6.1TAPWKK93 pKa = 10.3DD94 pKa = 3.47FHH96 pKa = 6.19FTEE99 pKa = 5.48IEE101 pKa = 3.92WTDD104 pKa = 3.07AGAALLPDD112 pKa = 4.84GVPGWLLGALGVLGVGAAASGGGGGGGGGGGPAFIPPLLPPQNRR156 pKa = 11.84GPEE159 pKa = 4.25GKK161 pKa = 9.84AEE163 pKa = 3.96PVATEE168 pKa = 3.49QDD170 pKa = 3.58KK171 pKa = 10.98PISGQVQATDD181 pKa = 3.77PDD183 pKa = 4.34RR184 pKa = 11.84DD185 pKa = 3.86TLNFTVAKK193 pKa = 9.39GPEE196 pKa = 4.27HH197 pKa = 5.83GTVTVDD203 pKa = 3.39PSTGQFLYY211 pKa = 10.18TPNPGYY217 pKa = 10.3EE218 pKa = 4.32GPDD221 pKa = 3.33SFEE224 pKa = 4.12VTVSDD229 pKa = 4.0GRR231 pKa = 11.84GGTTTVKK238 pKa = 10.54VPVTVSPVNDD248 pKa = 3.66APTAPDD254 pKa = 3.31YY255 pKa = 11.63TEE257 pKa = 4.2TTNEE261 pKa = 3.95DD262 pKa = 3.81TPVSGRR268 pKa = 11.84VTGSDD273 pKa = 3.2MDD275 pKa = 4.02SGDD278 pKa = 3.13TLTYY282 pKa = 11.07VKK284 pKa = 10.65GSDD287 pKa = 3.65PAHH290 pKa = 5.77GTVTVNPDD298 pKa = 2.63GTYY301 pKa = 10.05TYY303 pKa = 10.47TPNPDD308 pKa = 3.37FNGTDD313 pKa = 3.24SFTVTVSDD321 pKa = 3.49GHH323 pKa = 6.56GGTTTSTVTVTINPVNDD340 pKa = 3.1VPVFVDD346 pKa = 4.23GGGTPVNTTDD356 pKa = 4.08GYY358 pKa = 11.1VFGYY362 pKa = 10.61DD363 pKa = 3.27EE364 pKa = 4.79NRR366 pKa = 11.84PAGAVLGTVRR376 pKa = 11.84ATDD379 pKa = 3.22VDD381 pKa = 4.46SATVTYY387 pKa = 10.61SILSGNDD394 pKa = 2.77NGYY397 pKa = 9.62FAIDD401 pKa = 3.76PVTGEE406 pKa = 4.14ISLTPAGAAAFVNDD420 pKa = 3.89YY421 pKa = 10.49EE422 pKa = 5.65AGANAHH428 pKa = 5.79SLVVGANDD436 pKa = 3.74GTVTTSIPVTLNEE449 pKa = 4.18QNVNDD454 pKa = 4.63APTAPGDD461 pKa = 3.78TKK463 pKa = 9.37TTDD466 pKa = 2.82EE467 pKa = 4.47DD468 pKa = 4.64TPVSGQIVGTDD479 pKa = 3.11VDD481 pKa = 5.05GDD483 pKa = 3.72TLTYY487 pKa = 11.08VKK489 pKa = 10.65GSDD492 pKa = 3.65PAHH495 pKa = 5.68GTVTVNADD503 pKa = 2.83GTYY506 pKa = 9.88IYY508 pKa = 10.64TPGANFNGTDD518 pKa = 3.4SFTVTVSDD526 pKa = 3.49GHH528 pKa = 6.56GGTTTSTITVTVDD541 pKa = 3.27PVNDD545 pKa = 3.43APMVPDD551 pKa = 3.38YY552 pKa = 11.22TKK554 pKa = 8.55TTNEE558 pKa = 4.05DD559 pKa = 3.86TPVSGQVVGSDD570 pKa = 3.23VDD572 pKa = 4.34GDD574 pKa = 3.71TLTYY578 pKa = 11.02VKK580 pKa = 10.67GSDD583 pKa = 3.66PSHH586 pKa = 5.77GTVTVNADD594 pKa = 3.0GTYY597 pKa = 9.86TYY599 pKa = 11.45VPGANFNGTDD609 pKa = 3.4SFTVTVSDD617 pKa = 3.49GHH619 pKa = 6.56GGTTTSTVTVTINPVNDD636 pKa = 3.1VPVFVDD642 pKa = 4.23GGGTPVNTTDD652 pKa = 4.08GYY654 pKa = 11.1VFGYY658 pKa = 10.61DD659 pKa = 3.27EE660 pKa = 4.79NRR662 pKa = 11.84PAGAVLGTVRR672 pKa = 11.84ATDD675 pKa = 3.22VDD677 pKa = 4.46SATVTYY683 pKa = 10.61SILSGNDD690 pKa = 2.77NGYY693 pKa = 9.62FAIDD697 pKa = 3.76PVTGEE702 pKa = 4.14ISLTPAGAAAFVNDD716 pKa = 3.89YY717 pKa = 10.49EE718 pKa = 5.65AGANAHH724 pKa = 5.79SLVVGANDD732 pKa = 3.61GTVTTNIPVTLNEE745 pKa = 3.97QNINDD750 pKa = 4.58APTAPDD756 pKa = 3.34DD757 pKa = 3.95TKK759 pKa = 9.39TTNEE763 pKa = 4.07DD764 pKa = 3.84TPVSGQVTGSDD775 pKa = 3.34VDD777 pKa = 4.15GDD779 pKa = 3.91TLTYY783 pKa = 11.0AKK785 pKa = 10.34GSNPSHH791 pKa = 5.83GTVTVNADD799 pKa = 3.0GTYY802 pKa = 9.89TYY804 pKa = 11.45VPGPNFNGTDD814 pKa = 3.42SFTVTVSDD822 pKa = 3.49GHH824 pKa = 6.56GGTTTSTVNVTVNPVNDD841 pKa = 3.86APSATVNNAGVVSEE855 pKa = 4.45GSLPGGIPAAGEE867 pKa = 4.16PPLQSTGKK875 pKa = 10.21ISIADD880 pKa = 3.87PDD882 pKa = 4.0SAAGDD887 pKa = 3.8LSVSLSGPNGVTSGGQPVSWTWDD910 pKa = 3.26AGTHH914 pKa = 5.07TLTGSVTVGGVTTEE928 pKa = 3.83VMTVAVGNVTATGAGQFEE946 pKa = 4.58AGYY949 pKa = 8.04TVTLKK954 pKa = 11.09APIDD958 pKa = 3.73HH959 pKa = 6.93LPGNGEE965 pKa = 4.19GVSNLHH971 pKa = 6.29FEE973 pKa = 4.55AVVSDD978 pKa = 4.18GQASGAPVGFEE989 pKa = 4.27VPVKK993 pKa = 10.7DD994 pKa = 4.51DD995 pKa = 4.22APVLVNGEE1003 pKa = 4.07QAVDD1007 pKa = 3.48VAPIDD1012 pKa = 3.8TNLMVILDD1020 pKa = 4.33LSGSMGQEE1028 pKa = 3.64TPTRR1032 pKa = 11.84LSRR1035 pKa = 11.84AKK1037 pKa = 10.22EE1038 pKa = 4.12AIQNLIDD1045 pKa = 5.05GYY1047 pKa = 11.03DD1048 pKa = 3.12LYY1050 pKa = 11.8GDD1052 pKa = 3.68VRR1054 pKa = 11.84VQLVTFSTTGASQQAWMTAAEE1075 pKa = 4.42AKK1077 pKa = 10.75ALVQNLQASGSTNYY1091 pKa = 10.25DD1092 pKa = 3.03AALAAAMNGFSATGKK1107 pKa = 10.45LDD1109 pKa = 3.35GAQNVSYY1116 pKa = 10.17FLTDD1120 pKa = 3.7GEE1122 pKa = 4.4PTLGDD1127 pKa = 3.66GNTEE1131 pKa = 3.77QLANSSNSSTADD1143 pKa = 3.09RR1144 pKa = 11.84GIQAGEE1150 pKa = 3.57EE1151 pKa = 4.22AIWTNFLSTHH1161 pKa = 5.96QINSFALGLGSSLNAAAQAFIDD1183 pKa = 4.68PIAYY1187 pKa = 10.11NGNTGANTNGQIVTDD1202 pKa = 4.16TSQLNDD1208 pKa = 3.63MLQGTISVPPTVSNLLTGGLGGSSGFGADD1237 pKa = 3.36GGHH1240 pKa = 6.39VSTLSIDD1247 pKa = 3.45GTTYY1251 pKa = 11.35AFDD1254 pKa = 3.62SSTGLMTKK1262 pKa = 9.44TGPATGSDD1270 pKa = 3.75YY1271 pKa = 11.19SYY1273 pKa = 11.5NAATHH1278 pKa = 5.27QVTIATAQGGKK1289 pKa = 10.03LVVDD1293 pKa = 4.65FDD1295 pKa = 4.3SGEE1298 pKa = 3.84FSYY1301 pKa = 10.89QVAPRR1306 pKa = 11.84TSGSHH1311 pKa = 6.42NYY1313 pKa = 10.52DD1314 pKa = 3.02EE1315 pKa = 4.48TLSYY1319 pKa = 10.76QVVDD1323 pKa = 3.87RR1324 pKa = 11.84DD1325 pKa = 3.72GDD1327 pKa = 3.73ASNLATQTLHH1337 pKa = 6.57VNYY1340 pKa = 9.43TPAAGAAGAAALFSSADD1357 pKa = 3.6SLGLAGVDD1365 pKa = 3.33EE1366 pKa = 4.64ATVSLEE1372 pKa = 3.82QQQPQDD1378 pKa = 3.46APHH1381 pKa = 6.72FPFGDD1386 pKa = 4.12SPWGGTQLSDD1396 pKa = 3.38MLHH1399 pKa = 6.96AYY1401 pKa = 9.25NAAEE1405 pKa = 4.04SLQVLLQAAFPPAFASTQASPTVLALASSADD1436 pKa = 3.59SLTAYY1441 pKa = 9.61APPVAPSLDD1450 pKa = 4.22DD1451 pKa = 3.89EE1452 pKa = 4.71LHH1454 pKa = 6.08MGLALHH1460 pKa = 6.83HH1461 pKa = 6.68
MM1 pKa = 6.94EE2 pKa = 5.37AVYY5 pKa = 10.6KK6 pKa = 10.85SGGASRR12 pKa = 11.84KK13 pKa = 9.22AAADD17 pKa = 3.66TPLVLDD23 pKa = 4.13RR24 pKa = 11.84PSIVSLKK31 pKa = 10.04IAPEE35 pKa = 3.99SVLKK39 pKa = 9.89FEE41 pKa = 4.98RR42 pKa = 11.84RR43 pKa = 11.84GGDD46 pKa = 3.25LVLVLQGGQEE56 pKa = 3.97VAVRR60 pKa = 11.84GFFTEE65 pKa = 4.1YY66 pKa = 10.84SDD68 pKa = 4.27GGRR71 pKa = 11.84NDD73 pKa = 4.32LVLEE77 pKa = 4.93DD78 pKa = 4.05GAGVQWWGQYY88 pKa = 6.1TAPWKK93 pKa = 10.3DD94 pKa = 3.47FHH96 pKa = 6.19FTEE99 pKa = 5.48IEE101 pKa = 3.92WTDD104 pKa = 3.07AGAALLPDD112 pKa = 4.84GVPGWLLGALGVLGVGAAASGGGGGGGGGGGPAFIPPLLPPQNRR156 pKa = 11.84GPEE159 pKa = 4.25GKK161 pKa = 9.84AEE163 pKa = 3.96PVATEE168 pKa = 3.49QDD170 pKa = 3.58KK171 pKa = 10.98PISGQVQATDD181 pKa = 3.77PDD183 pKa = 4.34RR184 pKa = 11.84DD185 pKa = 3.86TLNFTVAKK193 pKa = 9.39GPEE196 pKa = 4.27HH197 pKa = 5.83GTVTVDD203 pKa = 3.39PSTGQFLYY211 pKa = 10.18TPNPGYY217 pKa = 10.3EE218 pKa = 4.32GPDD221 pKa = 3.33SFEE224 pKa = 4.12VTVSDD229 pKa = 4.0GRR231 pKa = 11.84GGTTTVKK238 pKa = 10.54VPVTVSPVNDD248 pKa = 3.66APTAPDD254 pKa = 3.31YY255 pKa = 11.63TEE257 pKa = 4.2TTNEE261 pKa = 3.95DD262 pKa = 3.81TPVSGRR268 pKa = 11.84VTGSDD273 pKa = 3.2MDD275 pKa = 4.02SGDD278 pKa = 3.13TLTYY282 pKa = 11.07VKK284 pKa = 10.65GSDD287 pKa = 3.65PAHH290 pKa = 5.77GTVTVNPDD298 pKa = 2.63GTYY301 pKa = 10.05TYY303 pKa = 10.47TPNPDD308 pKa = 3.37FNGTDD313 pKa = 3.24SFTVTVSDD321 pKa = 3.49GHH323 pKa = 6.56GGTTTSTVTVTINPVNDD340 pKa = 3.1VPVFVDD346 pKa = 4.23GGGTPVNTTDD356 pKa = 4.08GYY358 pKa = 11.1VFGYY362 pKa = 10.61DD363 pKa = 3.27EE364 pKa = 4.79NRR366 pKa = 11.84PAGAVLGTVRR376 pKa = 11.84ATDD379 pKa = 3.22VDD381 pKa = 4.46SATVTYY387 pKa = 10.61SILSGNDD394 pKa = 2.77NGYY397 pKa = 9.62FAIDD401 pKa = 3.76PVTGEE406 pKa = 4.14ISLTPAGAAAFVNDD420 pKa = 3.89YY421 pKa = 10.49EE422 pKa = 5.65AGANAHH428 pKa = 5.79SLVVGANDD436 pKa = 3.74GTVTTSIPVTLNEE449 pKa = 4.18QNVNDD454 pKa = 4.63APTAPGDD461 pKa = 3.78TKK463 pKa = 9.37TTDD466 pKa = 2.82EE467 pKa = 4.47DD468 pKa = 4.64TPVSGQIVGTDD479 pKa = 3.11VDD481 pKa = 5.05GDD483 pKa = 3.72TLTYY487 pKa = 11.08VKK489 pKa = 10.65GSDD492 pKa = 3.65PAHH495 pKa = 5.68GTVTVNADD503 pKa = 2.83GTYY506 pKa = 9.88IYY508 pKa = 10.64TPGANFNGTDD518 pKa = 3.4SFTVTVSDD526 pKa = 3.49GHH528 pKa = 6.56GGTTTSTITVTVDD541 pKa = 3.27PVNDD545 pKa = 3.43APMVPDD551 pKa = 3.38YY552 pKa = 11.22TKK554 pKa = 8.55TTNEE558 pKa = 4.05DD559 pKa = 3.86TPVSGQVVGSDD570 pKa = 3.23VDD572 pKa = 4.34GDD574 pKa = 3.71TLTYY578 pKa = 11.02VKK580 pKa = 10.67GSDD583 pKa = 3.66PSHH586 pKa = 5.77GTVTVNADD594 pKa = 3.0GTYY597 pKa = 9.86TYY599 pKa = 11.45VPGANFNGTDD609 pKa = 3.4SFTVTVSDD617 pKa = 3.49GHH619 pKa = 6.56GGTTTSTVTVTINPVNDD636 pKa = 3.1VPVFVDD642 pKa = 4.23GGGTPVNTTDD652 pKa = 4.08GYY654 pKa = 11.1VFGYY658 pKa = 10.61DD659 pKa = 3.27EE660 pKa = 4.79NRR662 pKa = 11.84PAGAVLGTVRR672 pKa = 11.84ATDD675 pKa = 3.22VDD677 pKa = 4.46SATVTYY683 pKa = 10.61SILSGNDD690 pKa = 2.77NGYY693 pKa = 9.62FAIDD697 pKa = 3.76PVTGEE702 pKa = 4.14ISLTPAGAAAFVNDD716 pKa = 3.89YY717 pKa = 10.49EE718 pKa = 5.65AGANAHH724 pKa = 5.79SLVVGANDD732 pKa = 3.61GTVTTNIPVTLNEE745 pKa = 3.97QNINDD750 pKa = 4.58APTAPDD756 pKa = 3.34DD757 pKa = 3.95TKK759 pKa = 9.39TTNEE763 pKa = 4.07DD764 pKa = 3.84TPVSGQVTGSDD775 pKa = 3.34VDD777 pKa = 4.15GDD779 pKa = 3.91TLTYY783 pKa = 11.0AKK785 pKa = 10.34GSNPSHH791 pKa = 5.83GTVTVNADD799 pKa = 3.0GTYY802 pKa = 9.89TYY804 pKa = 11.45VPGPNFNGTDD814 pKa = 3.42SFTVTVSDD822 pKa = 3.49GHH824 pKa = 6.56GGTTTSTVNVTVNPVNDD841 pKa = 3.86APSATVNNAGVVSEE855 pKa = 4.45GSLPGGIPAAGEE867 pKa = 4.16PPLQSTGKK875 pKa = 10.21ISIADD880 pKa = 3.87PDD882 pKa = 4.0SAAGDD887 pKa = 3.8LSVSLSGPNGVTSGGQPVSWTWDD910 pKa = 3.26AGTHH914 pKa = 5.07TLTGSVTVGGVTTEE928 pKa = 3.83VMTVAVGNVTATGAGQFEE946 pKa = 4.58AGYY949 pKa = 8.04TVTLKK954 pKa = 11.09APIDD958 pKa = 3.73HH959 pKa = 6.93LPGNGEE965 pKa = 4.19GVSNLHH971 pKa = 6.29FEE973 pKa = 4.55AVVSDD978 pKa = 4.18GQASGAPVGFEE989 pKa = 4.27VPVKK993 pKa = 10.7DD994 pKa = 4.51DD995 pKa = 4.22APVLVNGEE1003 pKa = 4.07QAVDD1007 pKa = 3.48VAPIDD1012 pKa = 3.8TNLMVILDD1020 pKa = 4.33LSGSMGQEE1028 pKa = 3.64TPTRR1032 pKa = 11.84LSRR1035 pKa = 11.84AKK1037 pKa = 10.22EE1038 pKa = 4.12AIQNLIDD1045 pKa = 5.05GYY1047 pKa = 11.03DD1048 pKa = 3.12LYY1050 pKa = 11.8GDD1052 pKa = 3.68VRR1054 pKa = 11.84VQLVTFSTTGASQQAWMTAAEE1075 pKa = 4.42AKK1077 pKa = 10.75ALVQNLQASGSTNYY1091 pKa = 10.25DD1092 pKa = 3.03AALAAAMNGFSATGKK1107 pKa = 10.45LDD1109 pKa = 3.35GAQNVSYY1116 pKa = 10.17FLTDD1120 pKa = 3.7GEE1122 pKa = 4.4PTLGDD1127 pKa = 3.66GNTEE1131 pKa = 3.77QLANSSNSSTADD1143 pKa = 3.09RR1144 pKa = 11.84GIQAGEE1150 pKa = 3.57EE1151 pKa = 4.22AIWTNFLSTHH1161 pKa = 5.96QINSFALGLGSSLNAAAQAFIDD1183 pKa = 4.68PIAYY1187 pKa = 10.11NGNTGANTNGQIVTDD1202 pKa = 4.16TSQLNDD1208 pKa = 3.63MLQGTISVPPTVSNLLTGGLGGSSGFGADD1237 pKa = 3.36GGHH1240 pKa = 6.39VSTLSIDD1247 pKa = 3.45GTTYY1251 pKa = 11.35AFDD1254 pKa = 3.62SSTGLMTKK1262 pKa = 9.44TGPATGSDD1270 pKa = 3.75YY1271 pKa = 11.19SYY1273 pKa = 11.5NAATHH1278 pKa = 5.27QVTIATAQGGKK1289 pKa = 10.03LVVDD1293 pKa = 4.65FDD1295 pKa = 4.3SGEE1298 pKa = 3.84FSYY1301 pKa = 10.89QVAPRR1306 pKa = 11.84TSGSHH1311 pKa = 6.42NYY1313 pKa = 10.52DD1314 pKa = 3.02EE1315 pKa = 4.48TLSYY1319 pKa = 10.76QVVDD1323 pKa = 3.87RR1324 pKa = 11.84DD1325 pKa = 3.72GDD1327 pKa = 3.73ASNLATQTLHH1337 pKa = 6.57VNYY1340 pKa = 9.43TPAAGAAGAAALFSSADD1357 pKa = 3.6SLGLAGVDD1365 pKa = 3.33EE1366 pKa = 4.64ATVSLEE1372 pKa = 3.82QQQPQDD1378 pKa = 3.46APHH1381 pKa = 6.72FPFGDD1386 pKa = 4.12SPWGGTQLSDD1396 pKa = 3.38MLHH1399 pKa = 6.96AYY1401 pKa = 9.25NAAEE1405 pKa = 4.04SLQVLLQAAFPPAFASTQASPTVLALASSADD1436 pKa = 3.59SLTAYY1441 pKa = 9.61APPVAPSLDD1450 pKa = 4.22DD1451 pKa = 3.89EE1452 pKa = 4.71LHH1454 pKa = 6.08MGLALHH1460 pKa = 6.83HH1461 pKa = 6.68
Molecular weight: 148.44 kDa
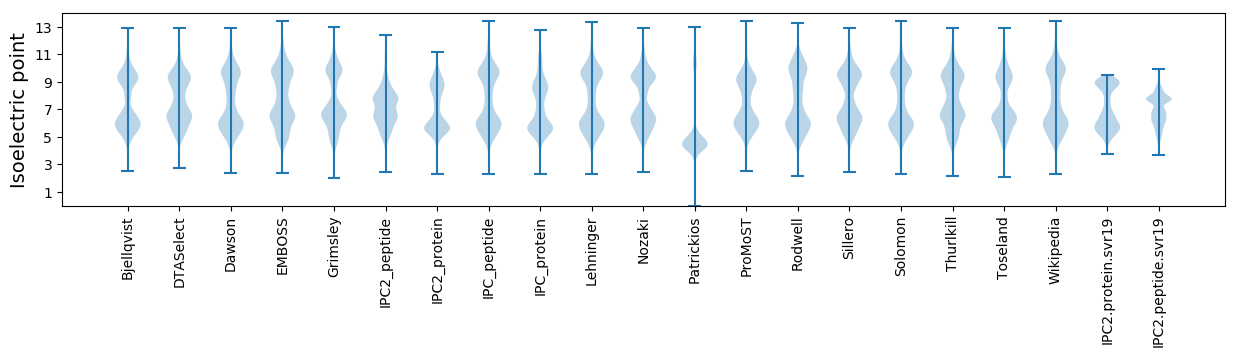
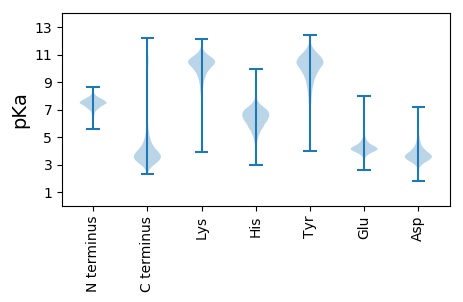
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H2M5N9|A0A0H2M5N9_VARPD Alpha-D-glucose-1-phosphatase OS=Variovorax paradoxus OX=34073 GN=VPARA_11970 PE=4 SV=1
MM1 pKa = 7.71PPSKK5 pKa = 10.42SPPSRR10 pKa = 11.84SPPSRR15 pKa = 11.84LPTRR19 pKa = 11.84PSSPNRR25 pKa = 11.84PPSKK29 pKa = 9.58PSSPSTPPSRR39 pKa = 11.84PSSPSRR45 pKa = 11.84LSTNPSSPSTPPSRR59 pKa = 11.84PSSARR64 pKa = 11.84RR65 pKa = 11.84LPTAPPLPSKK75 pKa = 10.12PSSPSRR81 pKa = 11.84PPTKK85 pKa = 9.62PSSPSTPPSRR95 pKa = 11.84PSSPSTPSVRR105 pKa = 11.84PPSRR109 pKa = 11.84LSSPPSNPPSGLSSSRR125 pKa = 11.84PLTAPSAPRR134 pKa = 11.84RR135 pKa = 11.84LSTRR139 pKa = 11.84PSWPSRR145 pKa = 11.84PPEE148 pKa = 4.4RR149 pKa = 11.84PSLPNKK155 pKa = 9.16PDD157 pKa = 3.6SAPSPPSRR165 pKa = 11.84PPTTPSPPSRR175 pKa = 11.84LPTGLPSPIRR185 pKa = 11.84PPSRR189 pKa = 11.84PSSPSTPPSTPPRR202 pKa = 11.84APSSPSRR209 pKa = 11.84PPTMPSSPITLSSRR223 pKa = 11.84LPMALPSPMRR233 pKa = 11.84PSSAPPSASKK243 pKa = 10.65PPTRR247 pKa = 11.84PSSPTSPARR256 pKa = 11.84RR257 pKa = 11.84LSWPSRR263 pKa = 11.84PSTTLPTTPSSPTRR277 pKa = 11.84PSRR280 pKa = 11.84APSRR284 pKa = 11.84PRR286 pKa = 11.84LPVRR290 pKa = 11.84PLTMPSSPTRR300 pKa = 11.84LSSPFCRR307 pKa = 11.84ASVTT311 pKa = 3.52
MM1 pKa = 7.71PPSKK5 pKa = 10.42SPPSRR10 pKa = 11.84SPPSRR15 pKa = 11.84LPTRR19 pKa = 11.84PSSPNRR25 pKa = 11.84PPSKK29 pKa = 9.58PSSPSTPPSRR39 pKa = 11.84PSSPSRR45 pKa = 11.84LSTNPSSPSTPPSRR59 pKa = 11.84PSSARR64 pKa = 11.84RR65 pKa = 11.84LPTAPPLPSKK75 pKa = 10.12PSSPSRR81 pKa = 11.84PPTKK85 pKa = 9.62PSSPSTPPSRR95 pKa = 11.84PSSPSTPSVRR105 pKa = 11.84PPSRR109 pKa = 11.84LSSPPSNPPSGLSSSRR125 pKa = 11.84PLTAPSAPRR134 pKa = 11.84RR135 pKa = 11.84LSTRR139 pKa = 11.84PSWPSRR145 pKa = 11.84PPEE148 pKa = 4.4RR149 pKa = 11.84PSLPNKK155 pKa = 9.16PDD157 pKa = 3.6SAPSPPSRR165 pKa = 11.84PPTTPSPPSRR175 pKa = 11.84LPTGLPSPIRR185 pKa = 11.84PPSRR189 pKa = 11.84PSSPSTPPSTPPRR202 pKa = 11.84APSSPSRR209 pKa = 11.84PPTMPSSPITLSSRR223 pKa = 11.84LPMALPSPMRR233 pKa = 11.84PSSAPPSASKK243 pKa = 10.65PPTRR247 pKa = 11.84PSSPTSPARR256 pKa = 11.84RR257 pKa = 11.84LSWPSRR263 pKa = 11.84PSTTLPTTPSSPTRR277 pKa = 11.84PSRR280 pKa = 11.84APSRR284 pKa = 11.84PRR286 pKa = 11.84LPVRR290 pKa = 11.84PLTMPSSPTRR300 pKa = 11.84LSSPFCRR307 pKa = 11.84ASVTT311 pKa = 3.52
Molecular weight: 32.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2201532 |
29 |
3308 |
324.3 |
35.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.322 ± 0.047 | 0.911 ± 0.01 |
5.096 ± 0.023 | 5.251 ± 0.026 |
3.552 ± 0.017 | 8.536 ± 0.03 |
2.221 ± 0.013 | 4.327 ± 0.019 |
3.178 ± 0.022 | 10.575 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.493 ± 0.016 | 2.516 ± 0.017 |
5.441 ± 0.022 | 3.69 ± 0.019 |
7.052 ± 0.029 | 5.448 ± 0.022 |
5.082 ± 0.022 | 7.688 ± 0.024 |
1.45 ± 0.012 | 2.17 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
