
Hyphomonas johnsonii MHS-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomonadales; Hyphomonadaceae; Hyphomonas; Hyphomonas johnsonii
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
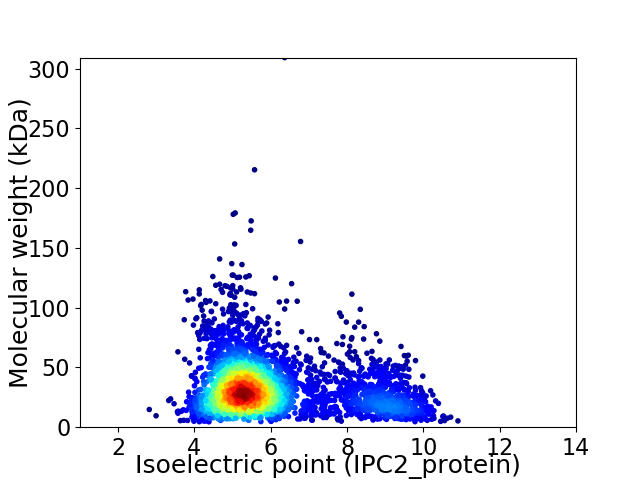
Virtual 2D-PAGE plot for 3447 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
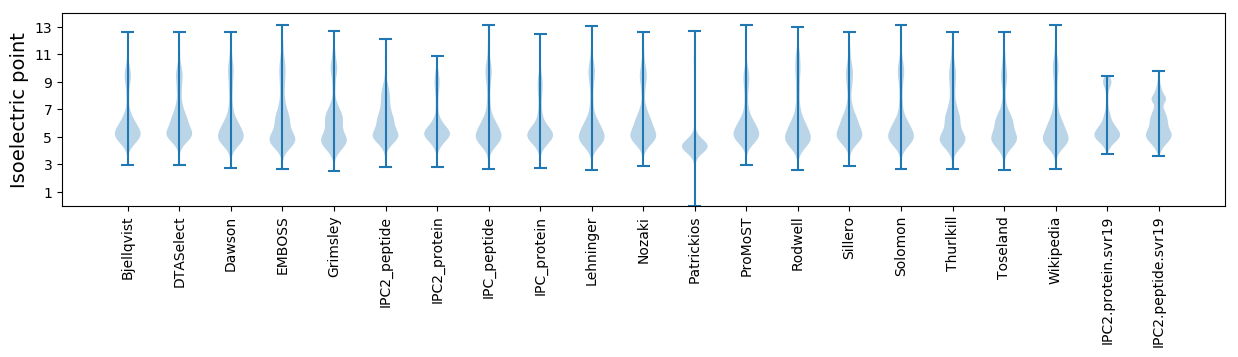
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A059FEC8|A0A059FEC8_9PROT Carboxyl transferase domain-containing protein OS=Hyphomonas johnsonii MHS-2 OX=1280950 GN=HJO_15189 PE=4 SV=1
MM1 pKa = 7.48NGNTIKK7 pKa = 10.73SRR9 pKa = 11.84LLASSVFAGVAFMASASMLATAQEE33 pKa = 4.45AEE35 pKa = 4.37DD36 pKa = 3.84TVDD39 pKa = 3.4TVEE42 pKa = 4.9APAAEE47 pKa = 4.64DD48 pKa = 3.14ATARR52 pKa = 11.84QATVYY57 pKa = 9.11VTGSRR62 pKa = 11.84IASPNLVTTSPVSTVTAADD81 pKa = 3.34IKK83 pKa = 10.99LQGATKK89 pKa = 10.5VEE91 pKa = 4.11DD92 pKa = 5.65LITQLPQAFAAQNSTVSNGASGTATVSLRR121 pKa = 11.84NLDD124 pKa = 3.81LGGSATRR131 pKa = 11.84TLVLIDD137 pKa = 4.35GKK139 pKa = 10.77RR140 pKa = 11.84MPYY143 pKa = 10.38GSPNDD148 pKa = 3.62AAADD152 pKa = 3.89LNLIPGALVEE162 pKa = 4.44RR163 pKa = 11.84VDD165 pKa = 3.85VLTGGASAVYY175 pKa = 10.33GSDD178 pKa = 2.82AVAGVVNFIMKK189 pKa = 10.55DD190 pKa = 3.16NFEE193 pKa = 4.27GVQLDD198 pKa = 3.57AQYY201 pKa = 11.75GFFQHH206 pKa = 6.53NNDD209 pKa = 3.21YY210 pKa = 8.77DD211 pKa = 4.05TNGNLRR217 pKa = 11.84SVIADD222 pKa = 4.23RR223 pKa = 11.84AATNPSQFQLPDD235 pKa = 3.77DD236 pKa = 4.15NVIDD240 pKa = 4.89GYY242 pKa = 10.9SKK244 pKa = 10.84EE245 pKa = 3.69ITAIMGVNTDD255 pKa = 4.01DD256 pKa = 4.52GRR258 pKa = 11.84GNLTGYY264 pKa = 10.37ISFRR268 pKa = 11.84DD269 pKa = 3.49NDD271 pKa = 3.6EE272 pKa = 4.07VLGGDD277 pKa = 3.88RR278 pKa = 11.84DD279 pKa = 3.94YY280 pKa = 11.39SACAIGSATATGFTCGGSATSVPGYY305 pKa = 10.59FLTQNNDD312 pKa = 2.69EE313 pKa = 4.44FTLDD317 pKa = 3.45SATGNTFRR325 pKa = 11.84PFDD328 pKa = 4.4SDD330 pKa = 3.26SDD332 pKa = 4.18LYY334 pKa = 11.75NFGPLNFYY342 pKa = 10.23QRR344 pKa = 11.84PDD346 pKa = 2.95QRR348 pKa = 11.84YY349 pKa = 7.9SVGFMGHH356 pKa = 5.95YY357 pKa = 9.27EE358 pKa = 4.02LNPNVEE364 pKa = 4.73AYY366 pKa = 7.74TQFMFMDD373 pKa = 4.07YY374 pKa = 10.8EE375 pKa = 4.32SRR377 pKa = 11.84AQIAPTGNFFSTNSINCDD395 pKa = 3.2NPLLSGSQVTDD406 pKa = 4.17LGCTPANVANGDD418 pKa = 4.01SVTMYY423 pKa = 9.32VARR426 pKa = 11.84RR427 pKa = 11.84NVEE430 pKa = 3.7GGGRR434 pKa = 11.84TDD436 pKa = 3.19NLGYY440 pKa = 8.05QTYY443 pKa = 9.97RR444 pKa = 11.84GVVGLRR450 pKa = 11.84GDD452 pKa = 3.97LFDD455 pKa = 4.49AAGWTYY461 pKa = 11.2DD462 pKa = 3.15VSAQYY467 pKa = 10.95SQVTLSRR474 pKa = 11.84AYY476 pKa = 8.96TNEE479 pKa = 3.49FSVTRR484 pKa = 11.84LNRR487 pKa = 11.84ALNVVDD493 pKa = 5.71DD494 pKa = 4.33GTGTPVCASVLDD506 pKa = 4.27GTDD509 pKa = 3.78PNCVPWDD516 pKa = 3.1IFTIGNVSQASLDD529 pKa = 3.88YY530 pKa = 11.01LQVPLLQNGTTTQNIVSAVVSGDD553 pKa = 2.98LGQYY557 pKa = 8.96GFKK560 pKa = 10.73SPAADD565 pKa = 2.93EE566 pKa = 4.44GMKK569 pKa = 10.15VAAGVEE575 pKa = 4.01YY576 pKa = 10.62RR577 pKa = 11.84RR578 pKa = 11.84DD579 pKa = 3.57SLEE582 pKa = 4.05SVTDD586 pKa = 3.52NSFATGDD593 pKa = 3.66GAGQGGPTIGLSGAIDD609 pKa = 3.5SYY611 pKa = 11.11EE612 pKa = 4.02AFGEE616 pKa = 4.21FQMPLVTGQPGVEE629 pKa = 3.98LLSIEE634 pKa = 4.03GAYY637 pKa = 9.94RR638 pKa = 11.84YY639 pKa = 10.03SDD641 pKa = 3.75YY642 pKa = 10.97STGSSSEE649 pKa = 4.08AYY651 pKa = 9.77KK652 pKa = 10.94LGGDD656 pKa = 3.77YY657 pKa = 11.18APTQDD662 pKa = 2.45IRR664 pKa = 11.84FRR666 pKa = 11.84ASYY669 pKa = 8.61QQAVRR674 pKa = 11.84APNVIDD680 pKa = 4.84LFQAQGFNLFDD691 pKa = 6.12LDD693 pKa = 6.14DD694 pKa = 4.94DD695 pKa = 4.9LCDD698 pKa = 3.95FTDD701 pKa = 4.31PEE703 pKa = 4.28NDD705 pKa = 3.53GTGGAACIGSNPWQVTQAQADD726 pKa = 3.96GGLLDD731 pKa = 5.0SPAGQYY737 pKa = 11.03NFLQGGNPNLEE748 pKa = 4.17PEE750 pKa = 4.17EE751 pKa = 4.61AKK753 pKa = 10.13TFTIGFVATPTFIPGLSLSLDD774 pKa = 3.78YY775 pKa = 11.5YY776 pKa = 11.24DD777 pKa = 5.42IDD779 pKa = 3.52IEE781 pKa = 4.33KK782 pKa = 10.68AISTVGASVTMQLCYY797 pKa = 10.85LEE799 pKa = 5.66GDD801 pKa = 3.49ADD803 pKa = 3.66ACSRR807 pKa = 11.84INRR810 pKa = 11.84NANGQLWVGTGNVVDD825 pKa = 3.88TNINIGGVSTSGYY838 pKa = 10.07DD839 pKa = 3.11VSAAYY844 pKa = 10.11GFDD847 pKa = 3.31AGTAGSFNLSMNGTYY862 pKa = 10.37LDD864 pKa = 4.34SFDD867 pKa = 4.4VDD869 pKa = 4.38PIGQSFAEE877 pKa = 4.11YY878 pKa = 10.47DD879 pKa = 3.79CVGEE883 pKa = 4.02YY884 pKa = 11.23GNDD887 pKa = 3.48CFTPTPEE894 pKa = 3.49WRR896 pKa = 11.84HH897 pKa = 5.15RR898 pKa = 11.84ARR900 pKa = 11.84LSWSTPVDD908 pKa = 3.49ALEE911 pKa = 5.46LNATWRR917 pKa = 11.84YY918 pKa = 9.15IGEE921 pKa = 4.12VDD923 pKa = 5.15LDD925 pKa = 3.67TGATGRR931 pKa = 11.84VDD933 pKa = 3.45STLDD937 pKa = 3.18AQNYY941 pKa = 8.89FDD943 pKa = 6.4LAGTWGATDD952 pKa = 3.41YY953 pKa = 10.39ATFRR957 pKa = 11.84FGVNNVLDD965 pKa = 4.89DD966 pKa = 5.01DD967 pKa = 5.07PPLSASVGTTGNNNTYY983 pKa = 8.6PQSYY987 pKa = 9.45DD988 pKa = 2.97ALGRR992 pKa = 11.84YY993 pKa = 8.95IFVGATLDD1001 pKa = 3.63FF1002 pKa = 4.87
MM1 pKa = 7.48NGNTIKK7 pKa = 10.73SRR9 pKa = 11.84LLASSVFAGVAFMASASMLATAQEE33 pKa = 4.45AEE35 pKa = 4.37DD36 pKa = 3.84TVDD39 pKa = 3.4TVEE42 pKa = 4.9APAAEE47 pKa = 4.64DD48 pKa = 3.14ATARR52 pKa = 11.84QATVYY57 pKa = 9.11VTGSRR62 pKa = 11.84IASPNLVTTSPVSTVTAADD81 pKa = 3.34IKK83 pKa = 10.99LQGATKK89 pKa = 10.5VEE91 pKa = 4.11DD92 pKa = 5.65LITQLPQAFAAQNSTVSNGASGTATVSLRR121 pKa = 11.84NLDD124 pKa = 3.81LGGSATRR131 pKa = 11.84TLVLIDD137 pKa = 4.35GKK139 pKa = 10.77RR140 pKa = 11.84MPYY143 pKa = 10.38GSPNDD148 pKa = 3.62AAADD152 pKa = 3.89LNLIPGALVEE162 pKa = 4.44RR163 pKa = 11.84VDD165 pKa = 3.85VLTGGASAVYY175 pKa = 10.33GSDD178 pKa = 2.82AVAGVVNFIMKK189 pKa = 10.55DD190 pKa = 3.16NFEE193 pKa = 4.27GVQLDD198 pKa = 3.57AQYY201 pKa = 11.75GFFQHH206 pKa = 6.53NNDD209 pKa = 3.21YY210 pKa = 8.77DD211 pKa = 4.05TNGNLRR217 pKa = 11.84SVIADD222 pKa = 4.23RR223 pKa = 11.84AATNPSQFQLPDD235 pKa = 3.77DD236 pKa = 4.15NVIDD240 pKa = 4.89GYY242 pKa = 10.9SKK244 pKa = 10.84EE245 pKa = 3.69ITAIMGVNTDD255 pKa = 4.01DD256 pKa = 4.52GRR258 pKa = 11.84GNLTGYY264 pKa = 10.37ISFRR268 pKa = 11.84DD269 pKa = 3.49NDD271 pKa = 3.6EE272 pKa = 4.07VLGGDD277 pKa = 3.88RR278 pKa = 11.84DD279 pKa = 3.94YY280 pKa = 11.39SACAIGSATATGFTCGGSATSVPGYY305 pKa = 10.59FLTQNNDD312 pKa = 2.69EE313 pKa = 4.44FTLDD317 pKa = 3.45SATGNTFRR325 pKa = 11.84PFDD328 pKa = 4.4SDD330 pKa = 3.26SDD332 pKa = 4.18LYY334 pKa = 11.75NFGPLNFYY342 pKa = 10.23QRR344 pKa = 11.84PDD346 pKa = 2.95QRR348 pKa = 11.84YY349 pKa = 7.9SVGFMGHH356 pKa = 5.95YY357 pKa = 9.27EE358 pKa = 4.02LNPNVEE364 pKa = 4.73AYY366 pKa = 7.74TQFMFMDD373 pKa = 4.07YY374 pKa = 10.8EE375 pKa = 4.32SRR377 pKa = 11.84AQIAPTGNFFSTNSINCDD395 pKa = 3.2NPLLSGSQVTDD406 pKa = 4.17LGCTPANVANGDD418 pKa = 4.01SVTMYY423 pKa = 9.32VARR426 pKa = 11.84RR427 pKa = 11.84NVEE430 pKa = 3.7GGGRR434 pKa = 11.84TDD436 pKa = 3.19NLGYY440 pKa = 8.05QTYY443 pKa = 9.97RR444 pKa = 11.84GVVGLRR450 pKa = 11.84GDD452 pKa = 3.97LFDD455 pKa = 4.49AAGWTYY461 pKa = 11.2DD462 pKa = 3.15VSAQYY467 pKa = 10.95SQVTLSRR474 pKa = 11.84AYY476 pKa = 8.96TNEE479 pKa = 3.49FSVTRR484 pKa = 11.84LNRR487 pKa = 11.84ALNVVDD493 pKa = 5.71DD494 pKa = 4.33GTGTPVCASVLDD506 pKa = 4.27GTDD509 pKa = 3.78PNCVPWDD516 pKa = 3.1IFTIGNVSQASLDD529 pKa = 3.88YY530 pKa = 11.01LQVPLLQNGTTTQNIVSAVVSGDD553 pKa = 2.98LGQYY557 pKa = 8.96GFKK560 pKa = 10.73SPAADD565 pKa = 2.93EE566 pKa = 4.44GMKK569 pKa = 10.15VAAGVEE575 pKa = 4.01YY576 pKa = 10.62RR577 pKa = 11.84RR578 pKa = 11.84DD579 pKa = 3.57SLEE582 pKa = 4.05SVTDD586 pKa = 3.52NSFATGDD593 pKa = 3.66GAGQGGPTIGLSGAIDD609 pKa = 3.5SYY611 pKa = 11.11EE612 pKa = 4.02AFGEE616 pKa = 4.21FQMPLVTGQPGVEE629 pKa = 3.98LLSIEE634 pKa = 4.03GAYY637 pKa = 9.94RR638 pKa = 11.84YY639 pKa = 10.03SDD641 pKa = 3.75YY642 pKa = 10.97STGSSSEE649 pKa = 4.08AYY651 pKa = 9.77KK652 pKa = 10.94LGGDD656 pKa = 3.77YY657 pKa = 11.18APTQDD662 pKa = 2.45IRR664 pKa = 11.84FRR666 pKa = 11.84ASYY669 pKa = 8.61QQAVRR674 pKa = 11.84APNVIDD680 pKa = 4.84LFQAQGFNLFDD691 pKa = 6.12LDD693 pKa = 6.14DD694 pKa = 4.94DD695 pKa = 4.9LCDD698 pKa = 3.95FTDD701 pKa = 4.31PEE703 pKa = 4.28NDD705 pKa = 3.53GTGGAACIGSNPWQVTQAQADD726 pKa = 3.96GGLLDD731 pKa = 5.0SPAGQYY737 pKa = 11.03NFLQGGNPNLEE748 pKa = 4.17PEE750 pKa = 4.17EE751 pKa = 4.61AKK753 pKa = 10.13TFTIGFVATPTFIPGLSLSLDD774 pKa = 3.78YY775 pKa = 11.5YY776 pKa = 11.24DD777 pKa = 5.42IDD779 pKa = 3.52IEE781 pKa = 4.33KK782 pKa = 10.68AISTVGASVTMQLCYY797 pKa = 10.85LEE799 pKa = 5.66GDD801 pKa = 3.49ADD803 pKa = 3.66ACSRR807 pKa = 11.84INRR810 pKa = 11.84NANGQLWVGTGNVVDD825 pKa = 3.88TNINIGGVSTSGYY838 pKa = 10.07DD839 pKa = 3.11VSAAYY844 pKa = 10.11GFDD847 pKa = 3.31AGTAGSFNLSMNGTYY862 pKa = 10.37LDD864 pKa = 4.34SFDD867 pKa = 4.4VDD869 pKa = 4.38PIGQSFAEE877 pKa = 4.11YY878 pKa = 10.47DD879 pKa = 3.79CVGEE883 pKa = 4.02YY884 pKa = 11.23GNDD887 pKa = 3.48CFTPTPEE894 pKa = 3.49WRR896 pKa = 11.84HH897 pKa = 5.15RR898 pKa = 11.84ARR900 pKa = 11.84LSWSTPVDD908 pKa = 3.49ALEE911 pKa = 5.46LNATWRR917 pKa = 11.84YY918 pKa = 9.15IGEE921 pKa = 4.12VDD923 pKa = 5.15LDD925 pKa = 3.67TGATGRR931 pKa = 11.84VDD933 pKa = 3.45STLDD937 pKa = 3.18AQNYY941 pKa = 8.89FDD943 pKa = 6.4LAGTWGATDD952 pKa = 3.41YY953 pKa = 10.39ATFRR957 pKa = 11.84FGVNNVLDD965 pKa = 4.89DD966 pKa = 5.01DD967 pKa = 5.07PPLSASVGTTGNNNTYY983 pKa = 8.6PQSYY987 pKa = 9.45DD988 pKa = 2.97ALGRR992 pKa = 11.84YY993 pKa = 8.95IFVGATLDD1001 pKa = 3.63FF1002 pKa = 4.87
Molecular weight: 106.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A059FNV0|A0A059FNV0_9PROT Pyruvate dehydrogenase E1 component subunit beta OS=Hyphomonas johnsonii MHS-2 OX=1280950 GN=HJO_08839 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.04RR14 pKa = 11.84THH16 pKa = 5.87GFRR19 pKa = 11.84LRR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84KK40 pKa = 8.16EE41 pKa = 3.63LSAA44 pKa = 4.99
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.04RR14 pKa = 11.84THH16 pKa = 5.87GFRR19 pKa = 11.84LRR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84KK40 pKa = 8.16EE41 pKa = 3.63LSAA44 pKa = 4.99
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1092738 |
40 |
2749 |
317.0 |
34.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.64 ± 0.062 | 0.848 ± 0.012 |
6.3 ± 0.032 | 5.711 ± 0.036 |
3.795 ± 0.025 | 8.628 ± 0.04 |
1.933 ± 0.02 | 5.121 ± 0.03 |
3.38 ± 0.035 | 9.655 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.612 ± 0.021 | 2.684 ± 0.027 |
5.147 ± 0.026 | 2.986 ± 0.02 |
6.57 ± 0.04 | 5.55 ± 0.03 |
5.537 ± 0.023 | 7.22 ± 0.029 |
1.383 ± 0.016 | 2.301 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
