
Rothia kristinae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Rothia
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
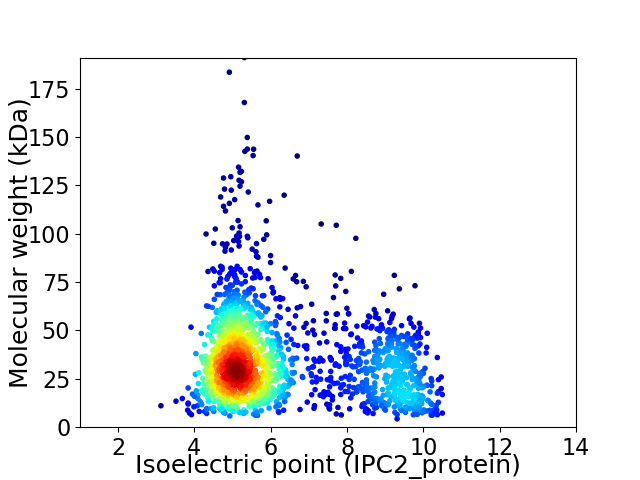
Virtual 2D-PAGE plot for 1872 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A199NUK7|A0A199NUK7_9MICC Thiol-disulfide isomerase OS=Rothia kristinae OX=37923 GN=AN277_0203740 PE=4 SV=1
MM1 pKa = 7.77ADD3 pKa = 3.44YY4 pKa = 11.36EE5 pKa = 4.34EE6 pKa = 4.89FDD8 pKa = 3.88GDD10 pKa = 3.68GEE12 pKa = 4.35EE13 pKa = 4.63YY14 pKa = 10.37EE15 pKa = 4.57RR16 pKa = 11.84EE17 pKa = 3.99APEE20 pKa = 3.97VYY22 pKa = 9.59RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84IIAAIIAILAIILVVWALVFAFQRR50 pKa = 11.84LTDD53 pKa = 4.02SGDD56 pKa = 3.36DD57 pKa = 3.34TAAASSAAAGDD68 pKa = 3.93NFDD71 pKa = 4.45SFSARR76 pKa = 11.84PTGGASGSASGSASAQPSASASAEE100 pKa = 3.77ATPTGEE106 pKa = 4.29STASADD112 pKa = 4.15AEE114 pKa = 4.34GTEE117 pKa = 4.77ASATPTDD124 pKa = 3.78QAAAPTPEE132 pKa = 4.58PTTQQPVQACSDD144 pKa = 3.62AFTVSTTVDD153 pKa = 3.33KK154 pKa = 10.96QVYY157 pKa = 9.52AAGQQPVITTTVANGSQNPCTVDD180 pKa = 3.23LGSANTTYY188 pKa = 10.77QITSGPANVYY198 pKa = 10.43SSQTCQAQPTHH209 pKa = 7.51DD210 pKa = 4.2EE211 pKa = 4.07ATLQAGAKK219 pKa = 7.42QTTSLTWDD227 pKa = 3.18RR228 pKa = 11.84GMNAYY233 pKa = 10.18GCGQAAQQAKK243 pKa = 8.35TGYY246 pKa = 8.57YY247 pKa = 9.04WVTATVNGVSSQPTLIVIQQ266 pKa = 3.9
MM1 pKa = 7.77ADD3 pKa = 3.44YY4 pKa = 11.36EE5 pKa = 4.34EE6 pKa = 4.89FDD8 pKa = 3.88GDD10 pKa = 3.68GEE12 pKa = 4.35EE13 pKa = 4.63YY14 pKa = 10.37EE15 pKa = 4.57RR16 pKa = 11.84EE17 pKa = 3.99APEE20 pKa = 3.97VYY22 pKa = 9.59RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84IIAAIIAILAIILVVWALVFAFQRR50 pKa = 11.84LTDD53 pKa = 4.02SGDD56 pKa = 3.36DD57 pKa = 3.34TAAASSAAAGDD68 pKa = 3.93NFDD71 pKa = 4.45SFSARR76 pKa = 11.84PTGGASGSASGSASAQPSASASAEE100 pKa = 3.77ATPTGEE106 pKa = 4.29STASADD112 pKa = 4.15AEE114 pKa = 4.34GTEE117 pKa = 4.77ASATPTDD124 pKa = 3.78QAAAPTPEE132 pKa = 4.58PTTQQPVQACSDD144 pKa = 3.62AFTVSTTVDD153 pKa = 3.33KK154 pKa = 10.96QVYY157 pKa = 9.52AAGQQPVITTTVANGSQNPCTVDD180 pKa = 3.23LGSANTTYY188 pKa = 10.77QITSGPANVYY198 pKa = 10.43SSQTCQAQPTHH209 pKa = 7.51DD210 pKa = 4.2EE211 pKa = 4.07ATLQAGAKK219 pKa = 7.42QTTSLTWDD227 pKa = 3.18RR228 pKa = 11.84GMNAYY233 pKa = 10.18GCGQAAQQAKK243 pKa = 8.35TGYY246 pKa = 8.57YY247 pKa = 9.04WVTATVNGVSSQPTLIVIQQ266 pKa = 3.9
Molecular weight: 27.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A199NTR6|A0A199NTR6_9MICC Uncharacterized protein OS=Rothia kristinae OX=37923 GN=AN277_0202340 PE=4 SV=1
MM1 pKa = 7.34SPRR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84SRR12 pKa = 11.84PRR14 pKa = 11.84PRR16 pKa = 11.84VRR18 pKa = 11.84MSTRR22 pKa = 11.84TRR24 pKa = 11.84FIVTVVAVVVLVVVGLGTRR43 pKa = 11.84GLDD46 pKa = 3.37LFRR49 pKa = 11.84GEE51 pKa = 5.25DD52 pKa = 3.63GSADD56 pKa = 3.61RR57 pKa = 11.84SGATASRR64 pKa = 11.84AAGPLPAGLTAQQALDD80 pKa = 3.6SLEE83 pKa = 4.44TKK85 pKa = 10.52GKK87 pKa = 9.83AARR90 pKa = 11.84TGYY93 pKa = 10.64SRR95 pKa = 11.84DD96 pKa = 3.28AFGARR101 pKa = 11.84WADD104 pKa = 3.35VDD106 pKa = 5.44HH107 pKa = 6.85NGCDD111 pKa = 3.32TRR113 pKa = 11.84NDD115 pKa = 2.97ILARR119 pKa = 11.84DD120 pKa = 4.17LTQTTLRR127 pKa = 11.84PGTCKK132 pKa = 10.37VATGVLQDD140 pKa = 4.2PYY142 pKa = 9.64TGRR145 pKa = 11.84RR146 pKa = 11.84IDD148 pKa = 3.75FTAGAQTSAAVQIDD162 pKa = 4.12HH163 pKa = 6.65VVALSDD169 pKa = 3.15AWQTGAQQLSTADD182 pKa = 3.82RR183 pKa = 11.84MALANDD189 pKa = 4.26PLNLLAVDD197 pKa = 4.99GPANNGKK204 pKa = 9.89SDD206 pKa = 3.55GDD208 pKa = 3.62AATWLPEE215 pKa = 4.0QKK217 pKa = 10.16SFRR220 pKa = 11.84CEE222 pKa = 3.58YY223 pKa = 9.74VARR226 pKa = 11.84QAGVKK231 pKa = 9.57RR232 pKa = 11.84KK233 pKa = 9.41YY234 pKa = 10.38DD235 pKa = 3.33LWVTRR240 pKa = 11.84AEE242 pKa = 4.0RR243 pKa = 11.84AAMQRR248 pKa = 11.84VLDD251 pKa = 3.88TCPGQRR257 pKa = 11.84IPTRR261 pKa = 3.58
MM1 pKa = 7.34SPRR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84SRR12 pKa = 11.84PRR14 pKa = 11.84PRR16 pKa = 11.84VRR18 pKa = 11.84MSTRR22 pKa = 11.84TRR24 pKa = 11.84FIVTVVAVVVLVVVGLGTRR43 pKa = 11.84GLDD46 pKa = 3.37LFRR49 pKa = 11.84GEE51 pKa = 5.25DD52 pKa = 3.63GSADD56 pKa = 3.61RR57 pKa = 11.84SGATASRR64 pKa = 11.84AAGPLPAGLTAQQALDD80 pKa = 3.6SLEE83 pKa = 4.44TKK85 pKa = 10.52GKK87 pKa = 9.83AARR90 pKa = 11.84TGYY93 pKa = 10.64SRR95 pKa = 11.84DD96 pKa = 3.28AFGARR101 pKa = 11.84WADD104 pKa = 3.35VDD106 pKa = 5.44HH107 pKa = 6.85NGCDD111 pKa = 3.32TRR113 pKa = 11.84NDD115 pKa = 2.97ILARR119 pKa = 11.84DD120 pKa = 4.17LTQTTLRR127 pKa = 11.84PGTCKK132 pKa = 10.37VATGVLQDD140 pKa = 4.2PYY142 pKa = 9.64TGRR145 pKa = 11.84RR146 pKa = 11.84IDD148 pKa = 3.75FTAGAQTSAAVQIDD162 pKa = 4.12HH163 pKa = 6.65VVALSDD169 pKa = 3.15AWQTGAQQLSTADD182 pKa = 3.82RR183 pKa = 11.84MALANDD189 pKa = 4.26PLNLLAVDD197 pKa = 4.99GPANNGKK204 pKa = 9.89SDD206 pKa = 3.55GDD208 pKa = 3.62AATWLPEE215 pKa = 4.0QKK217 pKa = 10.16SFRR220 pKa = 11.84CEE222 pKa = 3.58YY223 pKa = 9.74VARR226 pKa = 11.84QAGVKK231 pKa = 9.57RR232 pKa = 11.84KK233 pKa = 9.41YY234 pKa = 10.38DD235 pKa = 3.33LWVTRR240 pKa = 11.84AEE242 pKa = 4.0RR243 pKa = 11.84AAMQRR248 pKa = 11.84VLDD251 pKa = 3.88TCPGQRR257 pKa = 11.84IPTRR261 pKa = 3.58
Molecular weight: 28.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
632351 |
37 |
1820 |
337.8 |
36.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.238 ± 0.088 | 0.632 ± 0.014 |
5.7 ± 0.052 | 6.431 ± 0.064 |
2.843 ± 0.033 | 9.207 ± 0.057 |
2.151 ± 0.025 | 4.064 ± 0.044 |
2.071 ± 0.052 | 10.281 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.968 ± 0.024 | 1.742 ± 0.03 |
5.663 ± 0.048 | 3.437 ± 0.033 |
7.969 ± 0.071 | 5.261 ± 0.04 |
5.672 ± 0.039 | 8.238 ± 0.047 |
1.456 ± 0.022 | 1.975 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
